Different modes of retrovirus restriction by human APOBEC3A and APOBEC3G in vivo
- PMID: 24851906
- PMCID: PMC4031197
- DOI: 10.1371/journal.ppat.1004145
Different modes of retrovirus restriction by human APOBEC3A and APOBEC3G in vivo
Abstract
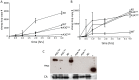
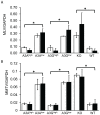
The apolipoprotein B editing complex 3 (A3) cytidine deaminases are among the most highly evolutionarily selected retroviral restriction factors, both in terms of gene copy number and sequence diversity. Primate genomes encode seven A3 genes, and while A3F and 3G are widely recognized as important in the restriction of HIV, the role of the other genes, particularly A3A, is not as clear. Indeed, since human cells can express multiple A3 genes, and because of the lack of an experimentally tractable model, it is difficult to dissect the individual contribution of each gene to virus restriction in vivo. To overcome this problem, we generated human A3A and A3G transgenic mice on a mouse A3 knockout background. Using these mice, we demonstrate that both A3A and A3G restrict infection by murine retroviruses but by different mechanisms: A3G was packaged into virions and caused extensive deamination of the retrovirus genomes while A3A was not packaged and instead restricted infection when expressed in target cells. Additionally, we show that a murine leukemia virus engineered to express HIV Vif overcame the A3G-mediated restriction, thereby creating a novel model for studying the interaction between these proteins. We have thus developed an in vivo system for understanding how human A3 proteins use different modes of restriction, as well as a means for testing therapies that disrupt HIV Vif-A3G interactions.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
-
- Ko TP, Lin JJ, Hu CY, Hsu YH, Wang AH, et al. (2003) Crystal structure of yeast cytosine deaminase. Insights into enzyme mechanism and evolution. J Biol Chem 278: 19111–19117. - PubMed
-
- Bishop KN, Holmes RK, Sheehy AM, Malim MH (2004) APOBEC-mediated editing of viral RNA. Science 305: 645–645. - PubMed
-
- Mangeat B, Turelli P, Caron G, Friedli M, Perrin L, et al. (2003) Broad antiretroviral defense by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424: 99–103. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

