RNA-Seq and expression microarray highlight different aspects of the fetal amniotic fluid transcriptome
- PMID: 24852236
- PMCID: PMC4184931
- DOI: 10.1002/pd.4417
RNA-Seq and expression microarray highlight different aspects of the fetal amniotic fluid transcriptome
Abstract
Objective: The aim of this study was to compare the complexity of the amniotic fluid supernatant cell-free fetal transcriptome as described by RNA Sequencing (RNA-Seq) and gene expression microarrays.
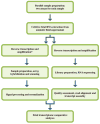
Methods: Cell-free fetal RNA from the amniotic fluid supernatant of five euploid mid-trimester samples was divided and prepared in tandem for analysis by either the Affymetrix HG-U133 Plus 2.0 Gene Chip microarray or Illumina HiSeq. Transcriptomes were assembled and compared on the basis of the presence of signal, rank-order gene expression, and pathway enrichment using Ingenuity Pathway Analysis (IPA). RNA-Seq data were also examined for evidence of alternative splicing.
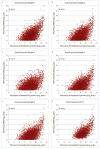
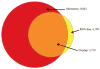
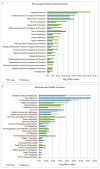
Results: Within individual samples, gene expression was strongly correlated (R = 0.43-0.57). Fewer expressed genes were observed using RNA-Seq than gene expression microarrays (4158 vs 8842). Most of the top pathways in the 'Physiological Systems Development and Function' IPA category were shared between platforms, although RNA-Seq yielded more significant p-values. Using RNA-Seq, examples of known alternative splicing were detected in several genes including H19 and IGF2.
Conclusions: In this pilot study, we found that expression microarrays gave a broader view of overall gene expression, while RNA-Seq demonstrated alternative splicing and specific pathways relevant to the developing fetus. The degraded nature of cell-free fetal RNA presented technical challenges for the RNA-Seq approach.
© 2014 John Wiley & Sons, Ltd.
Figures
References
-
- Larrabee PB, Johnson KL, Lai C, et al. Global gene expression analysis in the living human fetus using amniotic fluid: a feasibility study. JAMA. 2005;293:836–42. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous