Identification of a large protein network involved in epigenetic transmission in replicating DNA of embryonic stem cells
- PMID: 24852249
- PMCID: PMC4066787
- DOI: 10.1093/nar/gku374
Identification of a large protein network involved in epigenetic transmission in replicating DNA of embryonic stem cells
Abstract
Pluripotency of embryonic stem cells (ESCs) is maintained by transcriptional activities and chromatin modifying complexes highly organized within the chromatin. Although much effort has been focused on identifying genome-binding sites, little is known on their dynamic association with chromatin across cell divisions. Here, we used a modified version of the iPOND (isolation of proteins at nascent DNA) technology to identify a large protein network enriched at nascent DNA in ESCs. This comprehensive and unbiased proteomic characterization in ESCs reveals that, in addition to the core replication machinery, proteins relevant for pluripotency of ESCs are present at DNA replication sites. In particular, we show that the chromatin remodeller HDAC1-NuRD complex is enriched at nascent DNA. Interestingly, an acute block of HDAC1 in ESCs leads to increased acetylation of histone H3 lysine 9 at nascent DNA together with a concomitant loss of methylation. Consistently, in contrast to what has been described in tumour cell lines, these chromatin marks were found to be stable during cell cycle progression of ESCs. Our results are therefore compatible with a rapid deacetylation-coupled methylation mechanism during the replication of DNA in ESCs that may participate in the preservation of pluripotency of ESCs during replication.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

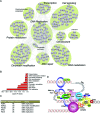
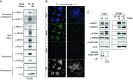

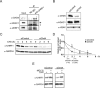

Similar articles
-
HDAC1 regulates pluripotency and lineage specific transcriptional networks in embryonic and trophoblast stem cells.Nucleic Acids Res. 2012 Apr;40(7):2925-39. doi: 10.1093/nar/gkr1151. Epub 2011 Dec 10. Nucleic Acids Res. 2012. PMID: 22156375 Free PMC article.
-
Ctbp2 Modulates NuRD-Mediated Deacetylation of H3K27 and Facilitates PRC2-Mediated H3K27me3 in Active Embryonic Stem Cell Genes During Exit from Pluripotency.Stem Cells. 2015 Aug;33(8):2442-55. doi: 10.1002/stem.2046. Epub 2015 May 26. Stem Cells. 2015. PMID: 25944056
-
An epigenetic switch regulates de novo DNA methylation at a subset of pluripotency gene enhancers during embryonic stem cell differentiation.Nucleic Acids Res. 2016 Sep 19;44(16):7605-17. doi: 10.1093/nar/gkw426. Epub 2016 May 13. Nucleic Acids Res. 2016. PMID: 27179026 Free PMC article.
-
Histone variants as emerging regulators of embryonic stem cell identity.Epigenetics. 2015;10(7):563-73. doi: 10.1080/15592294.2015.1053682. Epigenetics. 2015. PMID: 26114724 Free PMC article. Review.
-
The nucleosome remodeling and deacetylase complex in development and disease.Transl Res. 2015 Jan;165(1):36-47. doi: 10.1016/j.trsl.2014.05.003. Epub 2014 May 10. Transl Res. 2015. PMID: 24880148 Free PMC article. Review.
Cited by
-
DRUGPATH - a novel bioinformatic approach identifies DNA-damage pathway as a regulator of size maintenance in human ESCs and iPSCs.Sci Rep. 2019 Feb 13;9(1):1897. doi: 10.1038/s41598-018-37491-w. Sci Rep. 2019. PMID: 30760778 Free PMC article.
-
An emerging paradigm in epigenetic marking: coordination of transcription and replication.Transcription. 2024 Feb-Apr;15(1-2):22-37. doi: 10.1080/21541264.2024.2316965. Epub 2024 Feb 20. Transcription. 2024. PMID: 38378467 Free PMC article. Review.
-
Minichromosome Maintenance Proteins Cooperate with LANA during the G1/S Phase of the Cell Cycle To Support Viral DNA Replication.J Virol. 2019 Mar 21;93(7):e02256-18. doi: 10.1128/JVI.02256-18. Print 2019 Apr 1. J Virol. 2019. PMID: 30651368 Free PMC article.
-
Wnt/Tcf1 pathway restricts embryonic stem cell cycle through activation of the Ink4/Arf locus.PLoS Genet. 2017 Mar 27;13(3):e1006682. doi: 10.1371/journal.pgen.1006682. eCollection 2017 Mar. PLoS Genet. 2017. PMID: 28346462 Free PMC article.
-
Timing of Expansion of Fragile X Premutation Alleles During Intergenerational Transmission in a Mouse Model of the Fragile X-Related Disorders.Front Genet. 2018 Aug 10;9:314. doi: 10.3389/fgene.2018.00314. eCollection 2018. Front Genet. 2018. PMID: 30147707 Free PMC article.
References
-
- Tsubouchi T., Fisher A.G. Reprogramming and the pluripotent stem cell cycle. Curr. Top. Dev. Biol. 2013;104:223–241. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

