The deubiquitinase USP15 antagonizes Parkin-mediated mitochondrial ubiquitination and mitophagy
- PMID: 24852371
- PMCID: PMC7108632
- DOI: 10.1093/hmg/ddu244
The deubiquitinase USP15 antagonizes Parkin-mediated mitochondrial ubiquitination and mitophagy
Abstract
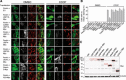
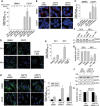
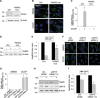
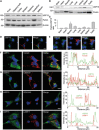
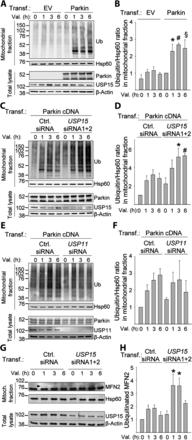
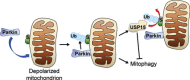
Loss-of-function mutations in PARK2, the gene encoding the E3 ubiquitin ligase Parkin, are the most frequent cause of recessive Parkinson's disease (PD). Parkin translocates from the cytosol to depolarized mitochondria, ubiquitinates outer mitochondrial membrane proteins and induces selective autophagy of the damaged mitochondria (mitophagy). Here, we show that ubiquitin-specific protease 15 (USP15), a deubiquitinating enzyme (DUB) widely expressed in brain and other organs, opposes Parkin-mediated mitophagy, while a panel of other DUBs and a catalytically inactive version of USP15 do not. Moreover, knockdown of USP15 rescues the mitophagy defect of PD patient fibroblasts with PARK2 mutations and decreased Parkin levels. USP15 does not affect the ubiquitination status of Parkin or Parkin translocation to mitochondria, but counteracts Parkin-mediated mitochondrial ubiquitination. Knockdown of the DUB CG8334, the closest homolog of USP15 in Drosophila, largely rescues the mitochondrial and behavioral defects of parkin RNAi flies. These data identify USP15 as an antagonist of Parkin and suggest that USP15 inhibition could be a therapeutic strategy for PD cases caused by reduced Parkin levels.
© The Author 2014. Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures


References
-
- Lees A.J., Hardy J., Revesz T. Parkinson's disease. Lancet. 2009;373:2055–2066. - PubMed
-
- Corti O., Lesage A., Brice A. What genetics tells us about the causes and mechanisms of Parkinson's disease. Physiol. Rev. 2011;91:1161–1218. - PubMed
-
- Pawlyk A.C., Giasson B.I., Sampathu D.M., Perez F.A., Lim K.L., Dawson V.L., Dawson T.M., Palmiter R.D., Trojanowski J.Q., Lee V.M. Novel monoclonal antibodies demonstrate biochemical variation of brain parkin with age. J. Biol. Chem. 2003;278:48120–48128. - PubMed
-
- Winklhofer K.F., Henn I.H., Kay-Jackson P.C., Heller U., Tatzelt J. Inactivation of parkin by oxidative stress and C-terminal truncations: a protective role of molecular chaperones. J. Biol. Chem. 2003;278:47199–47208. - PubMed
-
- Chung K.K., Thomas B., Li X., Pletnikova O., Troncoso J.C., Marsh L., Dawson V.L., Dawson T.M. S-nitrosylation of parkin regulates ubiquitination and compromises parkin's protective function. Science. 2004;304:1328–1331. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

