Multi-colour FISH in oesophageal adenocarcinoma-predictors of prognosis independent of stage and grade
- PMID: 24853183
- PMCID: PMC4056055
- DOI: 10.1038/bjc.2014.238
Multi-colour FISH in oesophageal adenocarcinoma-predictors of prognosis independent of stage and grade
Abstract
Background: Oesophageal adenocarcinoma or Barrett's adenocarcinoma (EAC) is increasing in incidence and stratification of prognosis might improve disease management. Multi-colour fluorescence in situ hybridisation (FISH) investigating ERBB2, MYC, CDKN2A and ZNF217 has recently shown promising results for the diagnosis of dysplasia and cancer using cytological samples.
Methods: To identify markers of prognosis we targeted four selected gene loci using multi-colour FISH applied to a tissue microarray containing 130 EAC samples. Prognostic predictors (P1, P2, P3) based on genomic copy numbers of the four loci were statistically assessed to stratify patients according to overall survival in combination with clinical data.
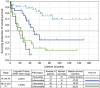
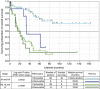
Results: The best stratification into favourable and unfavourable prognoses was shown by P1, percentage of cells with less than two ZNF217 signals; P2, percentage of cells with fewer ERBB2- than ZNF217 signals; and P3, overall ratio of ERBB2-/ZNF217 signals. Median survival times for P1 were 32 vs 73 months, 28 vs 73 months for P2; and 27 vs 65 months for P3. Regarding each tumour grade P2 subdivided patients into distinct prognostic groups independently within each grade, with different median survival times of at least 35 months.
Conclusions: Cell signal number of the ERBB2 and ZNF217 loci showed independence from tumour stage and differentiation grade. The prognostic value of multi-colour FISH-assays is applicable to EAC and is superior to single markers.
Figures




Similar articles
-
Fluorescence in situ hybridization identifies high risk Barrett's patients likely to develop esophageal adenocarcinoma.Dis Esophagus. 2016 Aug;29(6):513-9. doi: 10.1111/dote.12372. Epub 2015 Jun 5. Dis Esophagus. 2016. PMID: 26043762 Free PMC article.
-
Gains and amplifications of c-myc, EGFR, and 20.q13 loci in the no dysplasia-dysplasia-adenocarcinoma sequence of Barrett's esophagus.Cancer Epidemiol Biomarkers Prev. 2008 Jun;17(6):1380-5. doi: 10.1158/1055-9965.EPI-07-2734. Cancer Epidemiol Biomarkers Prev. 2008. PMID: 18559552
-
Fluorescence in situ hybridization mapping of esophagectomy specimens from patients with Barrett's esophagus with high-grade dysplasia or adenocarcinoma.Hum Pathol. 2012 Feb;43(2):172-9. doi: 10.1016/j.humpath.2011.04.018. Epub 2011 Aug 4. Hum Pathol. 2012. PMID: 21820152
-
The molecular biology of esophageal adenocarcinoma.J Surg Oncol. 2005 Dec 1;92(3):169-90. doi: 10.1002/jso.20359. J Surg Oncol. 2005. PMID: 16299787 Review.
-
Genetic and Epigenetic Alterations in Barrett's Esophagus and Esophageal Adenocarcinoma.Gastroenterol Clin North Am. 2015 Jun;44(2):473-89. doi: 10.1016/j.gtc.2015.02.015. Epub 2015 Apr 1. Gastroenterol Clin North Am. 2015. PMID: 26021206 Free PMC article. Review.
Cited by
-
Somatic deletion of KDM1A/LSD1 gene is associated to advanced colorectal cancer stages.J Clin Pathol. 2020 Feb;73(2):107-111. doi: 10.1136/jclinpath-2019-206128. Epub 2019 Aug 30. J Clin Pathol. 2020. PMID: 31471467 Free PMC article.
-
MET and PTEN gene copy numbers and Ki-67 protein expression associate with pathologic complete response in ERBB2-positive breast carcinoma patients treated with neoadjuvant trastuzumab-based therapy.BMC Cancer. 2016 Aug 30;16(1):695. doi: 10.1186/s12885-016-2743-x. BMC Cancer. 2016. PMID: 27576528 Free PMC article.
-
Molecular Biomarkers Detected Using Fluorescence in situ Hybridization in a Filipino with Retinoblastoma.Acta Med Philipp. 2024 Jun 14;58(10):99-107. doi: 10.47895/amp.vi0.7666. eCollection 2024. Acta Med Philipp. 2024. PMID: 38939426 Free PMC article.
-
Novel Criteria for Intratumoral Budding with Prognostic Relevance for Colon Cancer and Its Histological Subtypes.Int J Mol Sci. 2021 Dec 3;22(23):13108. doi: 10.3390/ijms222313108. Int J Mol Sci. 2021. PMID: 34884913 Free PMC article.
-
ZNF217: An Oncogenic Transcription Factor and Potential Therapeutic Target for Multiple Human Cancers.Cancer Manag Res. 2024 Jan 18;16:49-62. doi: 10.2147/CMAR.S431135. eCollection 2024. Cancer Manag Res. 2024. PMID: 38259608 Free PMC article. Review.
References
-
- Aitken SJ, Thomas JS, Langdon SP, Harrison DJ, Faratian D. Quantitative analysis of changes in ER, PR and HER2 expression in primary breast cancer and paired nodal metastases. Ann Oncol. 2010;21:1254–1261. - PubMed
-
- Akagi T, Ito T, Kato M, Jin Z, Cheng Y, Kan T, Yamamoto G, Olaru A, Kawamata N, Boult J, Soukiasian HJ, Miller CW, Ogawa S, Meltzer SJ, Koeffler HP. Chromosomal abnormalities and novel disease-related regions in progression from Barrett's esophagus to oesophageal adenocarcinoma. Int J Cancer. 2009;125:2349–2359. - PMC - PubMed
-
- Albrecht B, Hausmann M, Zitzelsberger H, Stein H, Siewert JR, Hopt U, Langer R, Höfler H, Werner M, Walch A. Array-based comparative genomic hybridisation for the detection of DNA sequence copy number changes in Barrett's adenocarcinoma. J Pathol. 2004;203:780–788. - PubMed
-
- Bang Y-J, Van Cutsem E, Feyereislova A, Chung HC, Shen L, Sawaki A, Lordick F, Ohtsu A, Omuro Y, Satoh T, Aprile G, Kulikov E, Hill J, Lehle M, Rüschoff J, Kang Y-K. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet. 2010;376:687–697. - PubMed
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

