IRE1a constitutes a negative feedback loop with BMP2 and acts as a novel mediator in modulating osteogenic differentiation
- PMID: 24853417
- PMCID: PMC4047903
- DOI: 10.1038/cddis.2014.194
IRE1a constitutes a negative feedback loop with BMP2 and acts as a novel mediator in modulating osteogenic differentiation
Erratum in
-
IRE1a constitutes a negative feedback loop with BMP2 and acts as a novel mediator in modulating osteogenic differentiation.Cell Death Dis. 2015 Apr 16;6(4):e1722. doi: 10.1038/cddis.2015.99. Cell Death Dis. 2015. PMID: 25880089 Free PMC article. No abstract available.
Expression of concern in
-
Expression of Concern to IRE1a constitutes a negative feedback loop with BMP2 and acts as a novel mediator in modulating osteogenic differentiation.Cell Death Dis. 2018 Nov 9;9(11):1124. doi: 10.1038/s41419-018-1175-8. Cell Death Dis. 2018. PMID: 30413683 Free PMC article.
Abstract
Bone morphogenetic protein 2 (BMP2) is known to activate unfolded protein response (UPR) signaling molecules, such as BiP (IgH chain-binding protein), PERK (PKR-like ER-resistant kinase), and IRE1α. Inositol-requiring enzyme-1a (IRE1a), as one of three unfolded protein sensors in UPR signaling pathways, can be activated during ER stress. Granulin-epithelin precursor (GEP) is an autocrine growth factor that has been implicated in embryonic development, tissue repair, tumorigenesis, and inflammation. However, the influence on IRE1a in BMP2-induced osteoblast differentiation has not yet been elucidated. Herein we demonstrate that overexpression of IRE1a inhibits osteoblast differentiation, as revealed by reduced activity of alkaline phosphatase (ALP) and osteocalcin; however, knockdown of IRE1a via the RNAi approach stimulates osteoblastogenesis. Mechanistic studies revealed that the expression of IRE1a during osteoblast was a consequence of JunB transcription factor binding to several AP1 sequence (TGAG/CTCA) in the 5'-flanking regulatory region of the IRE1a gene, followed by transcription. In addition, GEP induces IRE1a expressions and this induction of IRE1a by GEP depends on JunB. Furthermore, IRE1a inhibition of GEP-induced osteoblastogenesis relies on JunB. Besides, GEP is required for IRE1a inhibition of BMP2-induced bone formation. Collectively, these findings demonstrate that IRE1a negatively regulates BMP2-induced osteoblast differentiation and this IRE1a inhibition effect depends on GEP growth factor. Thus, IRE1a, BMP2, GEP growth factor, and JunB transcription factor form a regulatory loop and act in concert in the course of osteoblastogenesis.
Figures
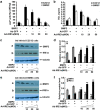




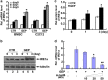



 ' indicate activation and repression, respectively
' indicate activation and repression, respectivelySimilar articles
-
Regulation of chondrocyte differentiation by IRE1α depends on its enzymatic activity.Cell Signal. 2014 Sep;26(9):1998-2007. doi: 10.1016/j.cellsig.2014.05.008. Epub 2014 May 23. Cell Signal. 2014. PMID: 24863879
-
IRE1α dissociates with BiP and inhibits ER stress-mediated apoptosis in cartilage development.Cell Signal. 2013 Nov;25(11):2136-46. doi: 10.1016/j.cellsig.2013.06.011. Epub 2013 Jun 29. Cell Signal. 2013. PMID: 23816533
-
XBP1S, a BMP2-inducible transcription factor, accelerates endochondral bone growth by activating GEP growth factor.J Cell Mol Med. 2014 Jun;18(6):1157-71. doi: 10.1111/jcmm.12261. Epub 2014 Mar 18. J Cell Mol Med. 2014. PMID: 24636354 Free PMC article.
-
Granulin epithelin precursor: a bone morphogenic protein 2-inducible growth factor that activates Erk1/2 signaling and JunB transcription factor in chondrogenesis.FASEB J. 2010 Jun;24(6):1879-92. doi: 10.1096/fj.09-144659. Epub 2010 Feb 2. FASEB J. 2010. PMID: 20124436 Free PMC article.
-
IRE1α pathway: A potential bone metabolism mediator.Cell Prolif. 2024 Oct;57(10):e13654. doi: 10.1111/cpr.13654. Epub 2024 May 12. Cell Prolif. 2024. PMID: 38736291 Free PMC article. Review.
Cited by
-
Extracellular vesicle-carried microRNA-27b derived from mesenchymal stem cells accelerates cutaneous wound healing via E3 ubiquitin ligase ITCH.J Cell Mol Med. 2020 Oct;24(19):11254-11271. doi: 10.1111/jcmm.15692. Epub 2020 Aug 26. J Cell Mol Med. 2020. PMID: 32845084 Free PMC article.
-
Expression of Concern to IRE1a constitutes a negative feedback loop with BMP2 and acts as a novel mediator in modulating osteogenic differentiation.Cell Death Dis. 2018 Nov 9;9(11):1124. doi: 10.1038/s41419-018-1175-8. Cell Death Dis. 2018. PMID: 30413683 Free PMC article.
-
Long Wavelength TCF-Based Fluorescent Probe for the Detection of Alkaline Phosphatase in Live Cells.Front Chem. 2019 Apr 30;7:255. doi: 10.3389/fchem.2019.00255. eCollection 2019. Front Chem. 2019. PMID: 31119120 Free PMC article.
-
Sphingosine Kinase 1 Protects Hepatocytes from Lipotoxicity via Down-regulation of IRE1α Protein Expression.J Biol Chem. 2015 Sep 18;290(38):23282-90. doi: 10.1074/jbc.M115.677542. Epub 2015 Aug 3. J Biol Chem. 2015. PMID: 26240153 Free PMC article.
-
The Role of Endoplasmic Reticulum Stress in Differentiation of Cells of Mesenchymal Origin.Biochemistry (Mosc). 2022 Sep;87(9):916-931. doi: 10.1134/S000629792209005X. Biochemistry (Mosc). 2022. PMID: 36180988 Free PMC article. Review.
References
-
- Lai CF, Cheng SL. Signal transductions induced by bone morphogenetic protein-2 and transforming growth factor-β in normal human osteoblastic cells. J Biol Chem. 2002;277:15514–15522. - PubMed
-
- Chen D, Zhao M, Mundy GR. Bone morphogenetic proteins. Growth Factors. 2004;22:233–241. - PubMed
-
- Murakami T, Saito A, Hino S, Kondo S, Kanemoto S, Chihara K, et al. Signaling mediated by the endoplasmic reticulum stress transducer OASIS is involved in bone formation. Nat Cell Biol. 2009;11:1205–1211. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

