Independent synchronized control and visualization of interactions between living cells and organisms
- PMID: 24853738
- PMCID: PMC4052360
- DOI: 10.1016/j.bpj.2014.03.044
Independent synchronized control and visualization of interactions between living cells and organisms
Abstract
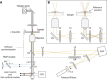
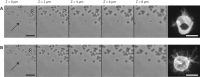
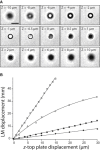
To investigate the early stages of cell-cell interactions occurring between living biological samples, imaging methods with appropriate spatiotemporal resolution are required. Among the techniques currently available, those based on optical trapping are promising. Methods to image trapped objects, however, in general suffer from a lack of three-dimensional resolution, due to technical constraints. Here, we have developed an original setup comprising two independent modules: holographic optical tweezers, which offer a versatile and precise way to move multiple objects simultaneously but independently, and a confocal microscope that provides fast three-dimensional image acquisition. The optical decoupling of these two modules through the same objective gives users the possibility to easily investigate very early steps in biological interactions. We illustrate the potential of this setup with an analysis of infection by the fungus Drechmeria coniospora of different developmental stages of Caenorhabditis elegans. This has allowed us to identify specific areas on the nematode's surface where fungal spores adhere preferentially. We also quantified this adhesion process for different mutant nematode strains, and thereby derive insights into the host factors that mediate fungal spore adhesion.
Copyright © 2014 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures




References
-
- Grakoui A., Bromley S.K., Dustin M.L. The immunological synapse: a molecular machine controlling T cell activation. Science. 1999;285:221–227. - PubMed
-
- Monks C.R., Freiberg B.A., Kupfer A. Three-dimensional segregation of supramolecular activation clusters in T cells. Nature. 1998;395:82–86. - PubMed
-
- Stuart L.M., Ezekowitz R.A. Phagocytosis and comparative innate immunity: learning on the fly. Nat. Rev. Immunol. 2008;8:131–141. - PubMed
-
- Wang Y., Botvinick E.L., Chien S. Visualizing the mechanical activation of Src. Nature. 2005;434:1040–1045. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

