A computational model for the loss of neuronal organization in microcolumns
- PMID: 24853752
- PMCID: PMC4052343
- DOI: 10.1016/j.bpj.2014.04.012
A computational model for the loss of neuronal organization in microcolumns
Abstract
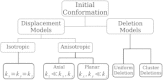
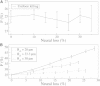
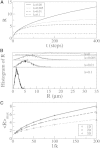
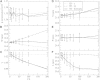
A population of neurons in the cerebral cortex of humans and other mammals organize themselves into vertical microcolumns perpendicular to the pial surface. Anatomical changes to these microcolumns have been correlated with neurological diseases and normal aging; in particular, in area 46 of the rhesus monkey brain, the strength of microcolumns was shown to decrease with age. These changes can be caused by alterations in the spatial distribution of the neurons in microcolumns and/or neuronal loss. Using a three-dimensional computational model of neuronal arrangements derived from thin tissue sections and validated in brain tissue from rhesus monkeys, we show that neuronal loss is inconsistent with the findings in aged individuals. In contrast, a model of simple random neuronal displacements, constrained in magnitude by restorative harmonic forces, is consistent with observed changes and provides mechanistic insights into the age-induced loss of microcolumnar structure. Connection of the model to normal aging and disease are discussed.
Copyright © 2014 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures


Similar articles
-
Age-related reduction in microcolumnar structure in area 46 of the rhesus monkey correlates with behavioral decline.Proc Natl Acad Sci U S A. 2004 Nov 9;101(45):15846-51. doi: 10.1073/pnas.0407002101. Epub 2004 Nov 1. Proc Natl Acad Sci U S A. 2004. PMID: 15520373 Free PMC article.
-
Generating a model of the three-dimensional spatial distribution of neurons using density maps.Neuroimage. 2008 Apr 15;40(3):1105-15. doi: 10.1016/j.neuroimage.2007.12.042. Epub 2008 Jan 5. Neuroimage. 2008. PMID: 18291677 Free PMC article.
-
A statistically based density map method for identification and quantification of regional differences in microcolumnarity in the monkey brain.J Neurosci Methods. 2005 Feb 15;141(2):321-32. doi: 10.1016/j.jneumeth.2004.09.005. J Neurosci Methods. 2005. PMID: 15661314
-
The Effects of Normal Aging on Nerve Fibers and Neuroglia in the Central Nervous System.In: Riddle DR, editor. Brain Aging: Models, Methods, and Mechanisms. Boca Raton (FL): CRC Press/Taylor & Francis; 2007. Chapter 5. In: Riddle DR, editor. Brain Aging: Models, Methods, and Mechanisms. Boca Raton (FL): CRC Press/Taylor & Francis; 2007. Chapter 5. PMID: 21204349 Free Books & Documents. Review.
-
The basic repeating modules of the cerebral cortical circuit.Proc Jpn Acad Ser B Phys Biol Sci. 2019;95(7):303-311. doi: 10.2183/pjab.95.022. Proc Jpn Acad Ser B Phys Biol Sci. 2019. PMID: 31406055 Free PMC article. Review.
Cited by
-
Contributions of Nonhuman Primates to Research on Aging.Vet Pathol. 2016 Mar;53(2):277-90. doi: 10.1177/0300985815622974. Epub 2016 Feb 11. Vet Pathol. 2016. PMID: 26869153 Free PMC article. Review.
References
-
- Mountcastle V.B. Modality and topographic properties of single neurons of cat’s somatic sensory cortex. J. Neurophysiol. 1957;20:408–434. - PubMed
-
- Mountcastle V.B. The columnar organization of the neocortex. Brain. 1997;120:701–722. - PubMed
-
- Buxhoeveden D.P., Casanova M.F. The minicolumn hypothesis in neuroscience. Brain. 2002;125:935–951. - PubMed
-
- Buxhoeveden D.P., Casanova M.F. The minicolumn and evolution of the brain. Brain Behav. Evol. 2002;60:125–151. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

