Loss-of-function variants in schizophrenia risk and SETD1A as a candidate susceptibility gene
- PMID: 24853937
- PMCID: PMC4387883
- DOI: 10.1016/j.neuron.2014.04.043
Loss-of-function variants in schizophrenia risk and SETD1A as a candidate susceptibility gene
Abstract
Loss-of-function (LOF) (i.e., nonsense, splice site, and frameshift) variants that lead to disruption of gene function are likely to contribute to the etiology of neuropsychiatric disorders. Here, we perform a systematic investigation of the role of both de novo and inherited LOF variants in schizophrenia using exome sequencing data from 231 case and 34 control trios. We identify two de novo LOF variants in the SETD1A gene, which encodes a subunit of histone methyltransferase, a finding unlikely to have occurred by chance, and provide evidence for a more general role of chromatin regulators in schizophrenia risk. Transmission pattern analyses reveal that LOF variants are more likely to be transmitted to affected individuals than controls. This is especially true for private LOF variants in genes intolerant to functional genetic variation. These findings highlight the contribution of LOF mutations to the genetic architecture of schizophrenia and provide important insights into disease pathogenesis.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures
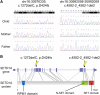
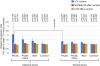
References
-
- Ben-David E, Shifman S. Combined analysis of exome sequencing points toward a major role for transcription regulation during brain development in autism. Mol. Psychiatry. 2013;18:1054–1056. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

