Targeted discovery and validation of plasma biomarkers of Parkinson's disease
- PMID: 24853996
- PMCID: PMC4224986
- DOI: 10.1021/pr500421v
Targeted discovery and validation of plasma biomarkers of Parkinson's disease
Abstract
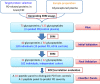
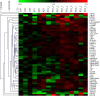
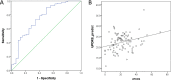
Despite extensive research, an unmet need remains for protein biomarkers of Parkinson's disease (PD) in peripheral body fluids, especially blood, which is easily accessible clinically. The discovery of such biomarkers is challenging, however, due to the enormous complexity and huge dynamic range of human blood proteins, which are derived from nearly all organ systems, with those originating specifically from the central nervous system (CNS) being exceptionally low in abundance. In this investigation of a relatively large cohort (∼300 subjects), selected reaction monitoring (SRM) assays (a targeted approach) were used to probe plasma peptides derived from glycoproteins previously found to be altered in the CNS based on PD diagnosis or severity. Next, the detected peptides were interrogated for their diagnostic sensitivity and specificity as well as the correlation with PD severity, as determined by the Unified Parkinson's Disease Rating Scale (UPDRS). The results revealed that 12 of the 50 candidate glycopeptides were reliably and consistently identified in plasma samples, with three of them displaying significant differences among diagnostic groups. A combination of four peptides (derived from PRNP, HSPG2, MEGF8, and NCAM1) provided an overall area under curve (AUC) of 0.753 (sensitivity: 90.4%; specificity: 50.0%). Additionally, combining two peptides (derived from MEGF8 and ICAM1) yielded significant correlation with PD severity, that is, UPDRS (r = 0.293, p = 0.004). The significance of these results is at least two-fold: (1) it is possible to use a targeted approach to identify otherwise very difficult to detect CNS related biomarkers in peripheral blood and (2) the novel biomarkers, if validated in independent cohorts, can be employed to assist with clinical diagnosis of PD as well as monitoring disease progression.
Keywords: Parkinson’s disease; peripheral biomarkers; selected reaction monitoring; targeted mass spectrometry.
Figures
Similar articles
-
Cerebrospinal fluid peptides as potential Parkinson disease biomarkers: a staged pipeline for discovery and validation.Mol Cell Proteomics. 2015 Mar;14(3):544-55. doi: 10.1074/mcp.M114.040576. Epub 2015 Jan 2. Mol Cell Proteomics. 2015. PMID: 25556233 Free PMC article.
-
Characterization of Parkinson's disease using blood-based biomarkers: A multicohort proteomic analysis.PLoS Med. 2019 Oct 11;16(10):e1002931. doi: 10.1371/journal.pmed.1002931. eCollection 2019 Oct. PLoS Med. 2019. PMID: 31603904 Free PMC article.
-
Serum microRNA expression levels in Turkish patients with Parkinson's disease.Int J Neurosci. 2021 Dec;131(12):1181-1189. doi: 10.1080/00207454.2020.1784165. Epub 2020 Jun 25. Int J Neurosci. 2021. PMID: 32546033
-
Mass spectrometry: A platform for biomarker discovery and validation for Alzheimer's and Parkinson's diseases.J Neurochem. 2019 Nov;151(4):397-416. doi: 10.1111/jnc.14635. Epub 2019 Jan 31. J Neurochem. 2019. PMID: 30474862 Free PMC article. Review.
-
Blood-based biomarkers for Parkinson's disease.Parkinsonism Relat Disord. 2014 Jan;20 Suppl 1(0 1):S99-103. doi: 10.1016/S1353-8020(13)70025-7. Parkinsonism Relat Disord. 2014. PMID: 24262199 Free PMC article. Review.
Cited by
-
Comparison of Ultraviolet Photodissociation and Collision Induced Dissociation of Adrenocorticotropic Hormone Peptides.J Am Soc Mass Spectrom. 2015 Sep;26(9):1570-9. doi: 10.1007/s13361-015-1186-y. Epub 2015 Jun 30. J Am Soc Mass Spectrom. 2015. PMID: 26122515
-
Gender specific decrease of a set of circulating N-acylphosphatidyl ethanolamines (NAPEs) in the plasma of Parkinson's disease patients.Metabolomics. 2019 May 3;15(5):74. doi: 10.1007/s11306-019-1536-z. Metabolomics. 2019. PMID: 31053995 Free PMC article.
-
Cerebrovascular-Specific Extracellular Matrix Bioink Promotes Blood-Brain Barrier Properties.Biomater Res. 2024 Dec 5;28:0115. doi: 10.34133/bmr.0115. eCollection 2024. Biomater Res. 2024. PMID: 39641002 Free PMC article.
-
Using Next-Generation Sequencing Transcriptomics To Determine Markers of Post-traumatic Symptoms: Preliminary Findings from a Post-deployment Cohort of Soldiers.G3 (Bethesda). 2019 Feb 7;9(2):463-471. doi: 10.1534/g3.118.200516. G3 (Bethesda). 2019. PMID: 30622122 Free PMC article.
-
Mapping human N-linked glycoproteins and glycosylation sites using mass spectrometry.Trends Analyt Chem. 2019 May;114:143-150. doi: 10.1016/j.trac.2019.02.009. Epub 2019 Feb 13. Trends Analyt Chem. 2019. PMID: 31831916 Free PMC article.
References
-
- Lang A. E.; Lozano A. M. Parkinson’s disease. First of two parts. N. Engl. J. Med. 1998, 339151044–1053. - PubMed
-
- Thomas B.; Beal M. F. Parkinson’s disease. Hum. Mol. Genet. 2007, 16Spec No. 2R183–194. - PubMed
-
- Shulman J. M.; De Jager P. L.; Feany M. B. Parkinson’s disease: genetics and pathogenesis. Annu. Rev. Pathol. 2011, 6, 193–222. - PubMed
-
- Jankovic J. Parkinson’s disease: clinical features and diagnosis. J. Neurol. Neurosurg. Psychiatry 2008, 794368–376. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- ES016873/ES/NIEHS NIH HHS/United States
- P42 ES004696/ES/NIEHS NIH HHS/United States
- P50 AG005131/AG/NIA NIH HHS/United States
- T32 ES015459/ES/NIEHS NIH HHS/United States
- P30 ES007033/ES/NIEHS NIH HHS/United States
- R01 AG033398/AG/NIA NIH HHS/United States
- ES004696-5897/ES/NIEHS NIH HHS/United States
- U01 NS082137/NS/NINDS NIH HHS/United States
- NS062684-6221/NS/NINDS NIH HHS/United States
- NS060252/NS/NINDS NIH HHS/United States
- AG033398/AG/NIA NIH HHS/United States
- R01 NS057567/NS/NINDS NIH HHS/United States
- NS062684/NS/NINDS NIH HHS/United States
- R43NS065553/NS/NINDS NIH HHS/United States
- NS082137/NS/NINDS NIH HHS/United States
- ES007033-6364/ES/NIEHS NIH HHS/United States
- NS057567/NS/NINDS NIH HHS/United States
- R43 NS065553/NS/NINDS NIH HHS/United States
- AGO5131/PHS HHS/United States
- R21 NS060252/NS/NINDS NIH HHS/United States
- R01 ES016873/ES/NIEHS NIH HHS/United States
- R01 ES019277/ES/NIEHS NIH HHS/United States
- P50 AG005136/AG/NIA NIH HHS/United States
- ES019277/ES/NIEHS NIH HHS/United States
- P50 NS062684/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

