Silencing of KIF14 interferes with cell cycle progression and cytokinesis by blocking the p27(Kip1) ubiquitination pathway in hepatocellular carcinoma
- PMID: 24854087
- PMCID: PMC4044675
- DOI: 10.1038/emm.2014.23
Silencing of KIF14 interferes with cell cycle progression and cytokinesis by blocking the p27(Kip1) ubiquitination pathway in hepatocellular carcinoma
Abstract
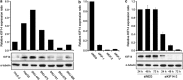
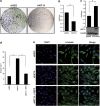
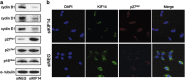
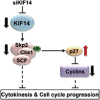
Although it has been suggested that kinesin family member 14 (KIF14) has oncogenic potential in various cancers, including hepatocellular carcinoma (HCC), the molecular mechanism of this potential remains unknown. We aimed to elucidate the role of KIF14 in hepatocarcinogenesis by knocking down KIF14 in HCC cells that overexpressed KIF14. After KIF14 knockdown, changes in tumor cell growth, cell cycle and cytokinesis were examined. We also examined cell cycle regulatory molecules and upstream Skp1/Cul1/F-box (SCF) complex molecules. Knockdown of KIF14 resulted in suppression of cell proliferation and failure of cytokinesis, whereas KIF14 overexpression increased cell proliferation. In KIF14-silenced cells, the levels of cyclins E1, D1 and B1 were profoundly decreased compared with control cells. Of the cyclin-dependent kinase inhibitors, the p27(Kip1) protein level specifically increased after KIF14 knockdown. The increase in p27(Kip1) was not due to elevation of its mRNA level, but was due to inhibition of the proteasome-dependent degradation pathway. To explore the pathway upstream of this event, we measured the levels of SCF complex molecules, including Skp1, Skp2, Cul1, Roc1 and Cks1. The levels of Skp2 and its cofactor Cks1 decreased in the KIF14 knockdown cells where p27(Kip1) accumulated. Overexpression of Skp2 in the KIF14 knockdown cells attenuated the failure of cytokinesis. On the basis of these results, we postulate that KIF14 knockdown downregulates the expression of Skp2 and Cks1, which target p27(Kip1) for degradation by the 26S proteasome, leading to accumulation of p27(Kip1). The downregulation of Skp2 and Cks1 also resulted in cytokinesis failure, which may inhibit tumor growth. To the best of our knowledge, this is the first report that has identified the molecular target and oncogenic effect of KIF14 in HCC.
Figures


References
-
- Corson TW, Huang A, Tsao MS, Gallie BL. KIF14 is a candidate oncogene in the 1q minimal region of genomic gain in multiple cancers. Oncogene. 2005;24:4741–4753. - PubMed
-
- Komatsu S, Imoto I, Tsuda H, Kozaki KI, Muramatsu T, Shimada Y, et al. Overexpression of SMYD2 relates to tumor cell proliferation and malignant outcome of esophageal squamous cell carcinoma. Carcinogenesis. 2009;30:1139–1146. - PubMed
-
- Roversi G, Pfundt R, Moroni RF, Magnani I, van Reijmersdal S, Pollo B, et al. Identification of novel genomic markers related to progression to glioblastoma through genomic profiling of 25 primary glioma cell lines. Oncogene. 2006;25:1571–1583. - PubMed
-
- Schlaeger C, Longerich T, Schiller C, Bewerunge P, Mehrabi A, Toedt G, et al. Etiology-dependent molecular mechanisms in human hepatocarcinogenesis. Hepatology. 2008;47:511–520. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous
