Deconstructing the peptide-MHC specificity of T cell recognition
- PMID: 24855945
- PMCID: PMC4071348
- DOI: 10.1016/j.cell.2014.03.047
Deconstructing the peptide-MHC specificity of T cell recognition
Abstract
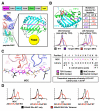
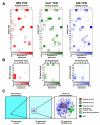
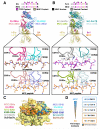
In order to survey a universe of major histocompatibility complex (MHC)-presented peptide antigens whose numbers greatly exceed the diversity of the T cell repertoire, T cell receptors (TCRs) are thought to be cross-reactive. However, the nature and extent of TCR cross-reactivity has not been conclusively measured experimentally. We developed a system to identify MHC-presented peptide ligands by combining TCR selection of highly diverse yeast-displayed peptide-MHC libraries with deep sequencing. Although we identified hundreds of peptides reactive with each of five different mouse and human TCRs, the selected peptides possessed TCR recognition motifs that bore a close resemblance to their known antigens. This structural conservation of the TCR interaction surface allowed us to exploit deep-sequencing information to computationally identify activating microbial and self-ligands for human autoimmune TCRs. The mechanistic basis of TCR cross-reactivity described here enables effective surveillance of diverse self and foreign antigens without necessitating degenerate recognition of nonhomologous peptides.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures




Comment in
-
Identifying environmental antigens that activate myelin-specific T cells.Trends Immunol. 2014 Jun;35(6):231-2. doi: 10.1016/j.it.2014.04.004. Epub 2014 May 9. Trends Immunol. 2014. PMID: 24820694 Free PMC article.
-
Focusing in on T cell cross-reactivity.Cell. 2014 May 22;157(5):1006-8. doi: 10.1016/j.cell.2014.05.002. Cell. 2014. PMID: 24855938
References
-
- Basu D, Horvath S, Matsumoto I, Fremont DH, Allen PM. Molecular basis for recognition of an arthritic peptide and a foreign epitope on distinct MHC molecules by a single TCR. J Immunol. 2000;164:5788–5796. - PubMed
-
- Benoist C, Mathis D. Autoimmunity provoked by infection: how good is the case for T cell epitope mimicry? Nat Immunol. 2001;2:797–801. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- SRA/SRP040021
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

