Opportunities and limitations of natural killer cells as adoptive therapy for malignant disease
- PMID: 24856895
- PMCID: PMC4190023
- DOI: 10.1016/j.jcyt.2014.03.009
Opportunities and limitations of natural killer cells as adoptive therapy for malignant disease
Abstract
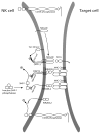
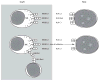
Although natural killer (NK) cells can be readily generated for adoptive therapy with current techniques, their optimal application to treat malignant diseases requires an appreciation of the dynamic balance between signals that either synergize with or antagonize each other. Individuals display wide differences in NK function that determine their therapeutic efficacy. The ability of NK cells to kill target cells or produce cytokines depends on the balance between signals from activating and inhibitory cell-surface receptors. The selection of NK cells with a predominant activating profile is critical for delivering successful anti-tumor activity. This can be achieved through selection of killer immunoglobulin-like receptor-mismatched NK donors and by use of blocking molecules against inhibitory pathways. Optimum NK cytotoxicity may require licensing or priming with tumor cells. Recent discoveries in the molecular and cellular biology of NK cells inform in the design of new strategies, including adjuvant therapies, to maximize the cytotoxic potential of NK cells for adoptive transfer to treat human malignancies.
Keywords: C-type lectin; graft-versus-leukemia; immunotherapy; killer immunoglobulin-like receptors; natural cytotoxicity receptors; natural killer.
Copyright © 2014 International Society for Cellular Therapy. All rights reserved.
Figures

References
-
- Lanier LL, Le AM, Civin CI, Loken MR, Phillips JH. The relationship of CD16 (Leu-11) and Leu-19 (NKH-1) antigen expression on human peripheral blood NK cells and cytotoxic T lymphocytes. J Immunol. 1986 Jun 15;136(12):4480–6. - PubMed
-
- Nagler A, Lanier LL, Cwirla S, Phillips JH. Comparative studies of human FcRIII-positive and negative natural killer cells. J Immunol. 1989 Nov 15;143(10):3183–91. - PubMed
-
- Cooper MA, Fehniger TA, Turner SC, Chen KS, Ghaheri BA, Ghayur T, et al. Human natural killer cells: a unique innate immunoregulatory role for the CD56(bright) subset. Blood. 2001 May 15;97(10):3146–51. - PubMed
-
- Campbell JJ, Qin S, Unutmaz D, Soler D, Murphy KE, Hodge MR, et al. Unique subpopulations of CD56+ NK and NK-T peripheral blood lymphocytes identified by chemokine receptor expression repertoire. J Immunol. 2001 Jun 1;166(11):6477–82. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

