Signaling hypergraphs
- PMID: 24857424
- PMCID: PMC4299695
- DOI: 10.1016/j.tibtech.2014.04.007
Signaling hypergraphs
Abstract
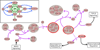
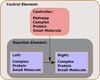
Signaling pathways function as the information-passing mechanisms of cells. A number of databases with extensive manual curation represent the current knowledge base for signaling pathways. These databases motivate the development of computational approaches for prediction and analysis. Such methods require an accurate and computable representation of signaling pathways. Pathways are often described as sets of proteins or as pairwise interactions between proteins. However, many signaling mechanisms cannot be described using these representations. In this opinion, we highlight a representation of signaling pathways that is underutilized: the hypergraph. We demonstrate the usefulness of hypergraphs in this context and discuss challenges and opportunities for the scientific community.
Keywords: graphs; hypergraphs; signaling pathways.
Copyright © 2014 Elsevier Ltd. All rights reserved.
Figures


References
-
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene Ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25(1):25–29. - PMC - PubMed
-
- Matthews L, Gopinath G, Gillespie M, Caudy M, Croft D, de Bono B, Garapati P, Hemish J, Hermjakob H, Jassal B, Kanapin A, Lewis S, Mahajan S, May B, Schmidt E, Vastrik I, Wu G, Birney E, Stein L, D’Eustachio P. Reactome knowledge-base of human biological pathways and processes. Nucleic Acids Research. 2009;37(Database issue):D619–D622. - PMC - PubMed
-
- Kandasamy K, Mohan SS, Raju R, Keerthikumar S, Kumar GS, Venugopal AK, Telikicherla D, Navarro JD, Mathivanan S, Pecquet C, Gollapudi SK, Tattikota SG, Mohan S, Padhukasahasram H, Subbannayya Y, Goel R, Jacob HK, Zhong J, Sekhar R, Nanjappa V, Balakrishnan L, Subbaiah R, Ramachandra YL, Rahiman BA, Prasad TS, Lin JX, Houtman JC, Desiderio S, Renauld JC, Constantinescu SN, Ohara O, Hirano T, Kubo M, Singh S, Khatri P, Draghici S, Bader GD, Sander C, Leonard WJ, Pandey A. NetPath: a public resource of curated signal transduction pathways. Genome Biology. 2010;11(1):R3. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

