Unambiguous identification of miRNA:target site interactions by different types of ligation reactions
- PMID: 24857550
- PMCID: PMC4181535
- DOI: 10.1016/j.molcel.2014.03.049
Unambiguous identification of miRNA:target site interactions by different types of ligation reactions
Abstract
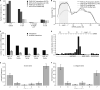
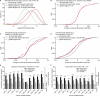
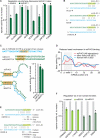
To exert regulatory function, miRNAs guide Argonaute (AGO) proteins to partially complementary sites on target RNAs. Crosslinking and immunoprecipitation (CLIP) assays are state-of-the-art to map AGO binding sites, but assigning the targeting miRNA to these sites relies on bioinformatics predictions and is therefore indirect. To directly and unambiguously identify miRNA:target site interactions, we modified our CLIP methodology in C. elegans to experimentally ligate miRNAs to their target sites. Unexpectedly, ligation reactions also occurred in the absence of the exogenous ligase. Our in vivo data set and reanalysis of published mammalian AGO-CLIP data for miRNA-chimeras yielded ∼17,000 miRNA:target site interactions. Analysis of interactions and extensive experimental validation of chimera-discovered targets of viral miRNAs suggest that our strategy identifies canonical, noncanonical, and nonconserved miRNA:targets. About 80% of miRNA interactions have perfect or partial seed complementarity. In summary, analysis of miRNA:target chimeras enables the systematic, context-specific, in vivo discovery of miRNA binding.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures

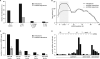
References
-
- Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, Eachus R, Pasquinelli AE. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
