An enhancer polymorphism at the cardiomyocyte intercalated disc protein NOS1AP locus is a major regulator of the QT interval
- PMID: 24857694
- PMCID: PMC4121472
- DOI: 10.1016/j.ajhg.2014.05.001
An enhancer polymorphism at the cardiomyocyte intercalated disc protein NOS1AP locus is a major regulator of the QT interval
Abstract
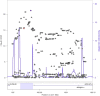
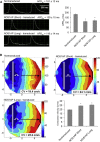
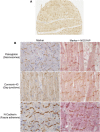
QT interval variation is assumed to arise from variation in repolarization as evidenced from rare Na- and K-channel mutations in Mendelian QT prolongation syndromes. However, in the general population, common noncoding variants at a chromosome 1q locus are the most common genetic regulators of QT interval variation. In this study, we use multiple human genetic, molecular genetic, and cellular assays to identify a functional variant underlying trait association: a noncoding polymorphism (rs7539120) that maps within an enhancer of NOS1AP and affects cardiac function by increasing NOS1AP transcript expression. We further localized NOS1AP to cardiomyocyte intercalated discs (IDs) and demonstrate that overexpression of NOS1AP in cardiomyocytes leads to altered cellular electrophysiology. We advance the hypothesis that NOS1AP affects cardiac electrical conductance and coupling and thereby regulates the QT interval through propagation defects. As further evidence of an important role for propagation variation affecting QT interval in humans, we show that common polymorphisms mapping near a specific set of 170 genes encoding ID proteins are significantly enriched for association with the QT interval, as compared to genome-wide markers. These results suggest that focused studies of proteins within the cardiomyocyte ID are likely to provide insights into QT prolongation and its associated disorders.
Copyright © 2014 The American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures


References
-
- Dekker J.M., Crow R.S., Hannan P.J., Schouten E.G., Folsom A.R., ARIC Study Heart rate-corrected QT interval prolongation predicts risk of coronary heart disease in black and white middle-aged men and women: the ARIC study. J. Am. Coll. Cardiol. 2004;43:565–571. - PubMed
-
- Newton-Cheh C., Larson M.G., Corey D.C., Benjamin E.J., Herbert A.G., Levy D., D’Agostino R.B., O’Donnell C.J. QT interval is a heritable quantitative trait with evidence of linkage to chromosome 3 in a genome-wide linkage analysis: The Framingham Heart Study. Heart Rhythm. 2005;2:277–284. - PubMed
-
- Priori S.G., Napolitano C. Genetics of cardiac arrhythmias and sudden cardiac death. Ann. N Y Acad. Sci. 2004;1015:96–110. - PubMed
-
- Arking D.E., Pfeufer A., Post W., Kao W.H., Newton-Cheh C., Ikeda M., West K., Kashuk C., Akyol M., Perz S. A common genetic variant in the NOS1 regulator NOS1AP modulates cardiac repolarization. Nat. Genet. 2006;38:644–651. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

