iHyd-PseAAC: predicting hydroxyproline and hydroxylysine in proteins by incorporating dipeptide position-specific propensity into pseudo amino acid composition
- PMID: 24857907
- PMCID: PMC4057693
- DOI: 10.3390/ijms15057594
iHyd-PseAAC: predicting hydroxyproline and hydroxylysine in proteins by incorporating dipeptide position-specific propensity into pseudo amino acid composition
Abstract
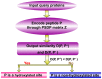
Post-translational modifications (PTMs) play crucial roles in various cell functions and biological processes. Protein hydroxylation is one type of PTM that usually occurs at the sites of proline and lysine. Given an uncharacterized protein sequence, which site of its Pro (or Lys) can be hydroxylated and which site cannot? This is a challenging problem, not only for in-depth understanding of the hydroxylation mechanism, but also for drug development, because protein hydroxylation is closely relevant to major diseases, such as stomach and lung cancers. With the avalanche of protein sequences generated in the post-genomic age, it is highly desired to develop computational methods to address this problem. In view of this, a new predictor called "iHyd-PseAAC" (identify hydroxylation by pseudo amino acid composition) was proposed by incorporating the dipeptide position-specific propensity into the general form of pseudo amino acid composition. It was demonstrated by rigorous cross-validation tests on stringent benchmark datasets that the new predictor is quite promising and may become a useful high throughput tool in this area. A user-friendly web-server for iHyd-PseAAC is accessible at http://app.aporc.org/iHyd-PseAAC/. Furthermore, for the convenience of the majority of experimental scientists, a step-by-step guide on how to use the web-server is given. Users can easily obtain their desired results by following these steps without the need of understanding the complicated mathematical equations presented in this paper just for its integrity.
Figures


Similar articles
-
iHyd-PseCp: Identify hydroxyproline and hydroxylysine in proteins by incorporating sequence-coupled effects into general PseAAC.Oncotarget. 2016 Jul 12;7(28):44310-44321. doi: 10.18632/oncotarget.10027. Oncotarget. 2016. PMID: 27322424 Free PMC article.
-
A Hybrid Deep Learning Model for Predicting Protein Hydroxylation Sites.Int J Mol Sci. 2018 Sep 18;19(9):2817. doi: 10.3390/ijms19092817. Int J Mol Sci. 2018. PMID: 30231550 Free PMC article.
-
iSNO-PseAAC: predict cysteine S-nitrosylation sites in proteins by incorporating position specific amino acid propensity into pseudo amino acid composition.PLoS One. 2013;8(2):e55844. doi: 10.1371/journal.pone.0055844. Epub 2013 Feb 7. PLoS One. 2013. PMID: 23409062 Free PMC article.
-
Recent Progress in Predicting Posttranslational Modification Sites in Proteins.Curr Top Med Chem. 2016;16(6):591-603. doi: 10.2174/1568026615666150819110421. Curr Top Med Chem. 2016. PMID: 26286211 Review.
-
pLoc_bal-mPlant: Predict Subcellular Localization of Plant Proteins by General PseAAC and Balancing Training Dataset.Curr Pharm Des. 2018;24(34):4013-4022. doi: 10.2174/1381612824666181119145030. Curr Pharm Des. 2018. PMID: 30451108 Review.
Cited by
-
Label-Free Method Development for Hydroxyproline PTM Mapping in Human Plasma Proteome.Protein J. 2021 Oct;40(5):741-755. doi: 10.1007/s10930-021-09984-7. Epub 2021 Apr 11. Protein J. 2021. PMID: 33840009
-
Impact of Bioinformatics Search Parameters for Peptides' Identification and Their Post-Translational Modifications: A Case Study of Proteolysed Gelatines from Beef, Pork, and Fish.Foods. 2023 Jun 28;12(13):2524. doi: 10.3390/foods12132524. Foods. 2023. PMID: 37444262 Free PMC article.
-
Detecting Succinylation sites from protein sequences using ensemble support vector machine.BMC Bioinformatics. 2018 Jun 25;19(1):237. doi: 10.1186/s12859-018-2249-4. BMC Bioinformatics. 2018. PMID: 29940836 Free PMC article.
-
iProt-Sub: a comprehensive package for accurately mapping and predicting protease-specific substrates and cleavage sites.Brief Bioinform. 2019 Mar 25;20(2):638-658. doi: 10.1093/bib/bby028. Brief Bioinform. 2019. PMID: 29897410 Free PMC article. Review.
-
iDNA-Prot|dis: identifying DNA-binding proteins by incorporating amino acid distance-pairs and reduced alphabet profile into the general pseudo amino acid composition.PLoS One. 2014 Sep 3;9(9):e106691. doi: 10.1371/journal.pone.0106691. eCollection 2014. PLoS One. 2014. PMID: 25184541 Free PMC article.
References
-
- Cockman M.E., Webb J.D., Kramer H.B., Kessler B.M., Ratcliffe P.J. Proteomics-based identification of novel factor inhibiting hypoxia-inducible factor (FIH) substrates indicates widespread asparaginyl hydroxylation of ankyrin repeat domain-containing proteins. Mol. Cell Proteomics. 2009;8:535–546. - PMC - PubMed
-
- Yamauchi M., Shiiba M. Lysine hydroxylation and cross-linking of collagen. Methods Mol. Biol. 2008;446:95–108. - PubMed
-
- Krane S.M. The importance of proline residues in the structure, stability and susceptibility to proteolytic degradation of collagens. Amino Acids. 2008;35:703–710. - PubMed
-
- Palfi V.K., Perczel A. How stable is a collagen triple helix? An ab initio study on various collagen and beta-sheet forming sequences. J. Comput. Chem. 2008;29:1374–1386. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

