Atomic force microscopy reveals a role for endothelial cell ICAM-1 expression in bladder cancer cell adherence
- PMID: 24857933
- PMCID: PMC4032264
- DOI: 10.1371/journal.pone.0098034
Atomic force microscopy reveals a role for endothelial cell ICAM-1 expression in bladder cancer cell adherence
Abstract
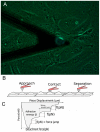
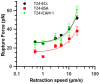
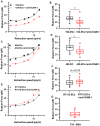
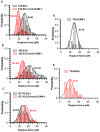
Cancer metastasis is a complex process involving cell-cell interactions mediated by cell adhesive molecules. In this study we determine the adhesion strength between an endothelial cell monolayer and tumor cells of different metastatic potentials using Atomic Force Microscopy. We show that the rupture forces of receptor-ligand bonds increase with retraction speed and range between 20 and 70 pN. It is shown that the most invasive cell lines (T24, J82) form the strongest bonds with endothelial cells. Using ICAM-1 coated substrates and a monoclonal antibody specific for ICAM-1, we demonstrate that ICAM-1 serves as a key receptor on endothelial cells and that its interactions with ligands expressed by tumor cells are correlated with the rupture forces obtained with the most invasive cancer cells (T24, J82). For the less invasive cancer cells (RT112), endothelial ICAM-1 does not seem to play any role in the adhesion process. Moreover, a detailed analysis of the distribution of rupture forces suggests that ICAM-1 interacts preferentially with one ligand on T24 cancer cells and with two ligands on J82 cancer cells. Possible counter receptors for these interactions are CD43 and MUC1, two known ligands for ICAM-1 which are expressed by these cancer cells.
Conflict of interest statement
Figures





References
-
- Miles FL, Pruitt FL, van Golen KL, Cooper CR (2008) Stepping out of the flow: capillary extravasation in cancer metastasis. Clin Exp Metastasis 25: 305–324. - PubMed
-
- Steeg PS (2006) Tumor metastasis: mechanistic insights and clinical challenges. Nat Med. 12: 895–904. - PubMed
-
- Haddad O, Chotard-Ghodsnia R, Verdier C, Duperray A (2010) Tumor cell/endothelial cell tight contact upregulates endothelial adhesion molecule expression mediated by NFkB: differential role of the shear stress. Exp. Cell Research 316: 615–626. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

