PTM-SD: a database of structurally resolved and annotated posttranslational modifications in proteins
- PMID: 24857970
- PMCID: PMC4038255
- DOI: 10.1093/database/bau041
PTM-SD: a database of structurally resolved and annotated posttranslational modifications in proteins
Abstract
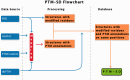
Posttranslational modifications (PTMs) define covalent and chemical modifications of protein residues. They play important roles in modulating various biological functions. Current PTM databases contain important sequence annotations but do not provide informative 3D structural resource about these modifications. Posttranslational modification structural database (PTM-SD) provides access to structurally solved modified residues, which are experimentally annotated as PTMs. It combines different PTM information and annotation gathered from other databases, e.g. Protein DataBank for the protein structures and dbPTM and PTMCuration for fine sequence annotation. PTM-SD gives an accurate detection of PTMs in structural data. PTM-SD can be browsed by PDB id, UniProt accession number, organism and classic PTM annotation. Advanced queries can also be performed, i.e. detailed PTM annotations, amino acid type, secondary structure, SCOP class classification, PDB chain length and number of PTMs by chain. Statistics and analyses can be computed on a selected dataset of PTMs. Each PTM entry is detailed in a dedicated page with information on the protein sequence, local conformation with secondary structure and Protein Blocks. PTM-SD gives valuable information on observed PTMs in protein 3D structure, which is of great interest for studying sequence-structure- function relationships at the light of PTMs, and could provide insights for comparative modeling and PTM predictions protocols. Database URL: PTM-SD can be accessed at http://www.dsimb.inserm.fr/dsimb_tools/PTM-SD/.
© The Author(s) 2014. Published by Oxford University Press.
Figures

Similar articles
-
dbPTM 2016: 10-year anniversary of a resource for post-translational modification of proteins.Nucleic Acids Res. 2016 Jan 4;44(D1):D435-46. doi: 10.1093/nar/gkv1240. Epub 2015 Nov 17. Nucleic Acids Res. 2016. PMID: 26578568 Free PMC article.
-
PRISMOID: a comprehensive 3D structure database for post-translational modifications and mutations with functional impact.Brief Bioinform. 2020 May 21;21(3):1069-1079. doi: 10.1093/bib/bbz050. Brief Bioinform. 2020. PMID: 31161204 Free PMC article.
-
dbPTM in 2022: an updated database for exploring regulatory networks and functional associations of protein post-translational modifications.Nucleic Acids Res. 2022 Jan 7;50(D1):D471-D479. doi: 10.1093/nar/gkab1017. Nucleic Acids Res. 2022. PMID: 34788852 Free PMC article.
-
Current status of PTMs structural databases: applications, limitations and prospects.Amino Acids. 2022 Apr;54(4):575-590. doi: 10.1007/s00726-021-03119-z. Epub 2022 Jan 12. Amino Acids. 2022. PMID: 35020020 Review.
-
The Methods Employed in Mass Spectrometric Analysis of Posttranslational Modifications (PTMs) and Protein-Protein Interactions (PPIs).Adv Exp Med Biol. 2019;1140:169-198. doi: 10.1007/978-3-030-15950-4_10. Adv Exp Med Biol. 2019. PMID: 31347048 Free PMC article. Review.
Cited by
-
Thirty years of molecular dynamics simulations on posttranslational modifications of proteins.Phys Chem Chem Phys. 2022 Nov 9;24(43):26371-26397. doi: 10.1039/d2cp02883b. Phys Chem Chem Phys. 2022. PMID: 36285789 Free PMC article. Review.
-
A curated rotamer library for common post-translational modifications of proteins.Bioinformatics. 2024 Jul 1;40(7):btae444. doi: 10.1093/bioinformatics/btae444. Bioinformatics. 2024. PMID: 38995731 Free PMC article.
-
Single-residue posttranslational modification sites at the N-terminus, C-terminus or in-between: To be or not to be exposed for enzyme access.Proteomics. 2015 Jul;15(14):2525-46. doi: 10.1002/pmic.201400633. Proteomics. 2015. PMID: 26038108 Free PMC article.
-
Structural Analysis of PTM Hotspots (SAPH-ire)--A Quantitative Informatics Method Enabling the Discovery of Novel Regulatory Elements in Protein Families.Mol Cell Proteomics. 2015 Aug;14(8):2285-97. doi: 10.1074/mcp.M115.051177. Epub 2015 Jun 12. Mol Cell Proteomics. 2015. PMID: 26070665 Free PMC article.
-
Role of Computational Methods in Going beyond X-ray Crystallography to Explore Protein Structure and Dynamics.Int J Mol Sci. 2018 Oct 30;19(11):3401. doi: 10.3390/ijms19113401. Int J Mol Sci. 2018. PMID: 30380757 Free PMC article. Review.
References
-
- Hubbard S.R., Till J.H. (2000) Protein tyrosine kinase structure and function. Annu. Rev. Biochem., 69, 373–398 http://www.ncbi.nlm.nih.gov/pubmed/10966463http://dx.doi.org/69/1/373 [pii] 10.1146/annurev.biochem.69.1.373 - PubMed
-
- Katre N.V., Wolber P.K., Stoeckenius W., Stroud R.M. (1981) Attachment site(s) of retinal in bacteriorhodopsin. Proc. Natl Acad. Sci. USA, 78, 4068–4072 http://www.ncbi.nlm.nih.gov/pubmed/6794028 - PMC - PubMed
-
- Anbalagan M., Huderson B., Murphy L., Rowan B.G. (2012) Post-translational modifications of nuclear receptors and human disease. Nucl. Recept. Signal., 10, e001 http://www.ncbi.nlm.nih.gov/pubmed/22438791http://dx.doi.org/10.1621/nrs.10001 - DOI - PMC - PubMed
-
- Oliveira A.P., Sauer U. (2012) The importance of post-translational modifications in regulating Saccharomyces cerevisiae metabolism. FEMS Yeast Res., 12, 104–117 http://www.ncbi.nlm.nih.gov/pubmed/22128902http://dx.doi.org/10.1111/j.1567-1364.2011.00765.x - DOI - PubMed
-
- Deribe Y.L., Pawson T., Dikic I. (2010) Post-translational modifications in signal integration. Nat. Struct. Mol. Biol., 17, 666–672 http://www.ncbi.nlm.nih.gov/pubmed/20495563http://dx.doi.org/nsmb.1842 [pii] 10.1038/nsmb.1842 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

