Integrated proteomics identified up-regulated focal adhesion-mediated proteins in human squamous cell carcinoma in an orthotopic murine model
- PMID: 24858105
- PMCID: PMC4032327
- DOI: 10.1371/journal.pone.0098208
Integrated proteomics identified up-regulated focal adhesion-mediated proteins in human squamous cell carcinoma in an orthotopic murine model
Abstract
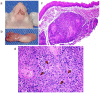
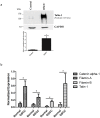
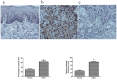
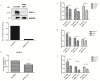
Understanding the molecular mechanisms of oral carcinogenesis will yield important advances in diagnostics, prognostics, effective treatment, and outcome of oral cancer. Hence, in this study we have investigated the proteomic and peptidomic profiles by combining an orthotopic murine model of oral squamous cell carcinoma (OSCC), mass spectrometry-based proteomics and biological network analysis. Our results indicated the up-regulation of proteins involved in actin cytoskeleton organization and cell-cell junction assembly events and their expression was validated in human OSCC tissues. In addition, the functional relevance of talin-1 in OSCC adhesion, migration and invasion was demonstrated. Taken together, this study identified specific processes deregulated in oral cancer and provided novel refined OSCC-targeting molecules.
Conflict of interest statement
Figures


References
-
- Siegel R, Naishadham D, Jemal A (2013) Cancer statistics, 2013. CA Cancer J Clin 63: 11–30. - PubMed
-
- Wong DT, Todd R, Tsuji T, Donoff RB (1996) Molecular biology of human oral cancer. Crit Rev Oral Biol Med 7: 319–328. - PubMed
-
- Warnakulasuriya S (2009) Global epidemiology of oral and oropharyngeal cancer. Oral Oncol 45: 309–316. - PubMed
-
- Choi S, Myers JN (2008) Molecular pathogenesis of oral squamous cell carcinoma: implications for therapy. J Dent Res 87: 14–32. - PubMed
-
- Panis C, Pizzatti L, Herrera AC, Cecchini R, Abdelhay E (2013) Putative circulating markers of the early and advanced stages of breast cancer identified by high-resolution label-free proteomics. Cancer Lett 330: 57–66. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

