MicroRNA-1 regulates chondrocyte phenotype by repressing histone deacetylase 4 during growth plate development
- PMID: 24858276
- PMCID: PMC4139910
- DOI: 10.1096/fj.13-249318
MicroRNA-1 regulates chondrocyte phenotype by repressing histone deacetylase 4 during growth plate development
Abstract
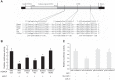
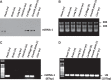
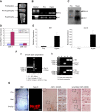
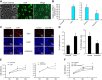
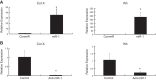
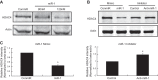
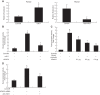
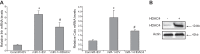
MicroRNAs (miRs) are noncoding RNAs (17-25 nt) that control translation and/or mRNA degradation. Using Northern blot analysis, we identified that miR-1 is specifically expressed in growth plate cartilage in addition to muscle tissue, but not in brain, intestine, liver, or lung. We obtained the first evidence that miR-1 is highly expressed in the hypertrophic zone of the growth plate, with an 8-fold increase compared with the proliferation zone; this location coincides with the Ihh and Col X expression regions in vivo. MiR-1 significantly induces chondrocyte proliferation and differentiation. We further identified histone deacetylase 4 (HDAC4) as a target of miR-1. HDAC4 negatively regulates chondrocyte hypertrophy by inhibiting Runx2, a critical transcription factor for chondrocyte hypertrophy. MiR-1 inhibits both endogenous HDAC4 protein by 2.2-fold and the activity of a reporter gene bearing the 3'-untranslated region (UTR) of HDAC4 by 3.3-fold. Conversely, knockdown of endogenous miR-1 relieves the repression of HDAC4. Deletion of the miR-1 binding site in HDAC4 3'-UTR or mutated miR-1 abolishes miR-1-mediated inhibition of the reporter gene activity. Overexpression of HDAC4 reverses miR-1 induction of chondrocyte differentiation markers Col X and Ihh. HDAC4 inhibits Runx2 promoter activity in a dosage-dependent manner. Thus, miR-1 plays an important role in the regulation of the chondrocyte phenotype during the growth plate development via direct targeting of HDAC4.
Keywords: HDAC4; Runx2; collagen X; differentiation.
© FASEB.
Figures
Similar articles
-
MiR-365: a mechanosensitive microRNA stimulates chondrocyte differentiation through targeting histone deacetylase 4.FASEB J. 2011 Dec;25(12):4457-66. doi: 10.1096/fj.11-185132. Epub 2011 Aug 19. FASEB J. 2011. PMID: 21856783 Free PMC article.
-
Subcellular relocation of histone deacetylase 4 regulates growth plate chondrocyte differentiation through Ca2+/calmodulin-dependent kinase IV.Am J Physiol Cell Physiol. 2012 Jul 1;303(1):C33-40. doi: 10.1152/ajpcell.00348.2011. Epub 2012 Mar 21. Am J Physiol Cell Physiol. 2012. PMID: 22442139 Free PMC article.
-
MicroRNA-381 Regulates Chondrocyte Hypertrophy by Inhibiting Histone Deacetylase 4 Expression.Int J Mol Sci. 2016 Aug 23;17(9):1377. doi: 10.3390/ijms17091377. Int J Mol Sci. 2016. PMID: 27563877 Free PMC article.
-
miRNA-based regulation in growth plate cartilage: mechanisms, targets, and therapeutic potential.Front Endocrinol (Lausanne). 2025 Mar 28;16:1530374. doi: 10.3389/fendo.2025.1530374. eCollection 2025. Front Endocrinol (Lausanne). 2025. PMID: 40225327 Free PMC article. Review.
-
HDAC4: an emerging target in diabetes mellitus and diabetic complications.Eur J Med Res. 2025 May 30;30(1):429. doi: 10.1186/s40001-025-02697-y. Eur J Med Res. 2025. PMID: 40448151 Free PMC article. Review.
Cited by
-
Compression regulates gene expression of chondrocytes through HDAC4 nuclear relocation via PP2A-dependent HDAC4 dephosphorylation.Biochim Biophys Acta. 2016 Jul;1863(7 Pt A):1633-42. doi: 10.1016/j.bbamcr.2016.04.018. Epub 2016 Apr 19. Biochim Biophys Acta. 2016. PMID: 27106144 Free PMC article.
-
miR-27b promotes type II collagen expression by targetting peroxisome proliferator-activated receptor-γ2 during rat articular chondrocyte differentiation.Biosci Rep. 2018 Jan 10;38(1):BSR20171109. doi: 10.1042/BSR20171109. Print 2018 Feb 28. Biosci Rep. 2018. PMID: 29187585 Free PMC article.
-
MicroRNA Contents in Matrix Vesicles Produced by Growth Plate Chondrocytes are Cell Maturation Dependent.Sci Rep. 2018 Feb 26;8(1):3609. doi: 10.1038/s41598-018-21517-4. Sci Rep. 2018. PMID: 29483516 Free PMC article.
-
Conditional deletion of HDAC4 from collagen type 2α1-expressing cells increases angiogenesis in vivo.Mol Med. 2020 May 1;26(1):36. doi: 10.1186/s10020-020-00154-6. Mol Med. 2020. PMID: 32354322 Free PMC article.
-
The role of histone deacetylase 4 during chondrocyte hypertrophy and endochondral bone development.Bone Joint Res. 2020 May 16;9(2):82-89. doi: 10.1302/2046-3758.92.BJR-2019-0172.R1. eCollection 2020 Feb. Bone Joint Res. 2020. PMID: 32435460 Free PMC article. Review.
References
-
- Redfern A. D., Colley S. M., Beveridge D. J., Ikeda N., Epis M. R., Li X., Foulds C. E., Stuart L. M., Barker A., Russell V. J., Ramsay K., Kobelke S. J., Li X., Hatchell E. C., Payne C., Giles K. M., Messineo A., Gatignol A., Lanz R. B., O'Malley B. W., Leedman P. J. (2013) RNA-induced silencing complex (RISC) proteins PACT, TRBP, and Dicer are SRA binding nuclear receptor coregulators. Proc. Natl. Acad. Sci. U. S. A. 110, 6536–6541 - PMC - PubMed
-
- Valencia-Sanchez M. A., Liu J., Hannon G. J., Parker R. (2006) Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 20, 515–524 - PubMed
-
- Bagga S., Bracht J., Hunter S., Massirer K., Holtz J., Eachus R., Pasquinelli A. E. (2005) Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell 122, 553–563 - PubMed
-
- Lee R. C., Ambros V. (2001) An extensive class of small RNAs in Caenorhabditis elegans. Science 294, 862–864 [See comment] - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

