Identification of recurrent SMO and BRAF mutations in ameloblastomas
- PMID: 24859340
- PMCID: PMC4418232
- DOI: 10.1038/ng.2986
Identification of recurrent SMO and BRAF mutations in ameloblastomas
Erratum in
- Nat Genet. 2015 Jan;47(1):97
Abstract
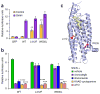
Here we report the discovery of oncogenic mutations in the Hedgehog and mitogen-activated protein kinase (MAPK) pathways in over 80% of ameloblastomas, locally destructive odontogenic tumors of the jaw, by genomic analysis of archival material. Mutations in SMO (encoding Smoothened, SMO) are common in ameloblastomas of the maxilla, whereas BRAF mutations are predominant in tumors of the mandible. We show that a frequently occurring SMO alteration encoding p.Leu412Phe is an activating mutation and that its effect on Hedgehog-pathway activity can be inhibited by arsenic trioxide (ATO), an anti-leukemia drug approved by the US Food and Drug Administration (FDA) that is currently in clinical trials for its Hedgehog-inhibitory activity. In a similar manner, ameloblastoma cells harboring an activating BRAF mutation encoding p.Val600Glu are sensitive to the BRAF inhibitor vemurafenib. Our findings establish a new paradigm for the diagnostic classification and treatment of ameloblastomas.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

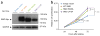

References
-
- Gorlin RJ, et al. Odontogenic tumors. Classification, histopathology, and clinical behavior in man and domesticated animals. Cancer. 1961;14:73–101. - PubMed
-
- Tucker A, et al. The cutting-edge of mammalian development; how the embryo makes teeth. Nat Rev Genet. 2004;5:499–508. - PubMed
-
- Davies H, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. - PubMed
-
- Parada LF, et al. Cooperation between gene encoding p53 tumour antigen and ras in cellular transformation. Nature. 1984;312:649–651. - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

