The molecular basis of ligand interaction at free fatty acid receptor 4 (FFA4/GPR120)
- PMID: 24860101
- PMCID: PMC4106347
- DOI: 10.1074/jbc.M114.561449
The molecular basis of ligand interaction at free fatty acid receptor 4 (FFA4/GPR120)
Abstract
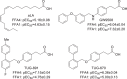
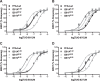
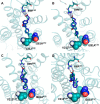
The long-chain fatty acid receptor FFA4 (previously GPR120) is receiving substantial interest as a novel target for the treatment of metabolic and inflammatory disease. This study examines for the first time the detailed mode of binding of both long-chain fatty acid and synthetic agonist ligands at FFA4 by integrating molecular modeling, receptor mutagenesis, and ligand structure-activity relationship approaches in an iterative format. In doing so, residues required for binding of fatty acid and synthetic agonists to FFA4 have been identified. This has allowed for the refinement of a well validated model of the mode of ligand-FFA4 interaction that will be invaluable in the identification of novel ligands and the future development of this receptor as a therapeutic target. The model reliably predicted the effects of substituent variations on agonist potency, and it was also able to predict the qualitative effect of binding site mutations in the majority of cases.
Keywords: 7-Helix Receptor; Bioluminescence Resonance Energy Transfer (BRET); Diabetes; Fatty Acid; G Protein-coupled Receptor (GPCR); Homology Modeling.
© 2014 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures





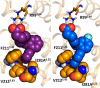
References
-
- Hirasawa A., Tsumaya K., Awaji T., Katsuma S., Adachi T., Yamada M., Sugimoto Y., Miyazaki S., Tsujimoto G. (2005) Free fatty acids regulate gut incretin glucagon-like peptide-1 secretion through GPR120. Nat. Med. 11, 90–94 - PubMed
-
- Engelstoft M. S., Park W.-M., Sakata I., Kristensen L. V., Husted A. S., Osborne-Lawrence S., Piper P. K., Walker A. K., Pedersen M. H., Nøhr M. K., Pan J., Sinz C. J., Carrington P. E., Akiyama T. E., Jones R. M., Tang C., Ahmed K., Offermanns S., Egerod K. L., Zigman J. M., Schwartz T. W. (2013) Seven transmembrane G protein-coupled receptor repertoire of gastric ghrelin cells. Mol. Metab. 2, 376–392 - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

