A forward genetic screen reveals essential and non-essential RNAi factors in Paramecium tetraurelia
- PMID: 24860163
- PMCID: PMC4066745
- DOI: 10.1093/nar/gku223
A forward genetic screen reveals essential and non-essential RNAi factors in Paramecium tetraurelia
Abstract
In most eukaryotes, small RNA-mediated gene silencing pathways form complex interacting networks. In the ciliate Paramecium tetraurelia, at least two RNA interference (RNAi) mechanisms coexist, involving distinct but overlapping sets of protein factors and producing different types of short interfering RNAs (siRNAs). One is specifically triggered by high-copy transgenes, and the other by feeding cells with double-stranded RNA (dsRNA)-producing bacteria. In this study, we designed a forward genetic screen for mutants deficient in dsRNA-induced silencing, and a powerful method to identify the relevant mutations by whole-genome sequencing. We present a set of 47 mutant alleles for five genes, revealing two previously unknown RNAi factors: a novel Paramecium-specific protein (Pds1) and a Cid1-like nucleotidyl transferase. Analyses of allelic diversity distinguish non-essential and essential genes and suggest that the screen is saturated for non-essential, single-copy genes. We show that non-essential genes are specifically involved in dsRNA-induced RNAi while essential ones are also involved in transgene-induced RNAi. One of the latter, the RNA-dependent RNA polymerase RDR2, is further shown to be required for all known types of siRNAs, as well as for sexual reproduction. These results open the way for the dissection of the genetic complexity, interconnection, mechanisms and natural functions of RNAi pathways in P. tetraurelia.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
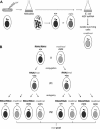


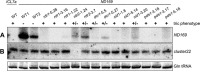
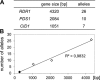
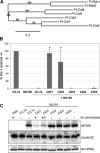

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

