Brd4 activates P-TEFb for RNA polymerase II CTD phosphorylation
- PMID: 24860166
- PMCID: PMC4081074
- DOI: 10.1093/nar/gku449
Brd4 activates P-TEFb for RNA polymerase II CTD phosphorylation
Abstract
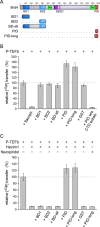
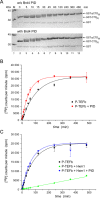
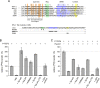
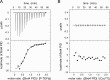
The bromodomain protein Brd4 regulates the transcription of signal-inducible genes. This is achieved by recruiting the positive transcription elongation factor P-TEFb to promoters by its P-TEFb interaction domain (PID). Here we show that Brd4 stimulates the kinase activity of P-TEFb for phosphorylation of the C-terminal domain (CTD) of RNA polymerase II over basal levels. The CTD phosphorylation saturation levels, the preferences for pre-phosphorylated substrates, and the phosphorylation specificity for Ser5 of the CTD however remain unchanged. Inhibition of P-TEFb by Hexim1 is relieved by Brd4, although no mutual displacement with the Cyclin T-binding domain of Hexim1 was observed. Brd4 PID shows a surprising sequence motif similarity to the trans-activating Tat protein from HIV-1, which includes a core RxL motif, a polybasic cluster known as arginine-rich motif, and a C-terminal leucine motif. Mutation of these motifs to alanine significantly diminished the stimulatory effect of Brd4 and fully abrogated its activation potential in presence of Hexim1. Yet the protein was not found to bind Cyclin T1 as Tat, but only P-TEFb with a dissociation constant of 0.5 μM. Our data suggest a model where Brd4 acts on the kinase subunit of P-TEFb to relieve inhibition and stimulate substrate recognition.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

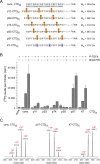
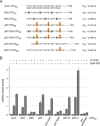

References
-
- Eick D., Geyer M. The RNA polymerase II carboxy-terminal domain (CTD) code. Chem. Rev. 2013;113:8456–8490. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

