Frizzled-9 impairs acetylcholine receptor clustering in skeletal muscle cells
- PMID: 24860427
- PMCID: PMC4029016
- DOI: 10.3389/fncel.2014.00110
Frizzled-9 impairs acetylcholine receptor clustering in skeletal muscle cells
Abstract
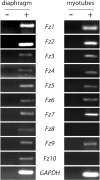
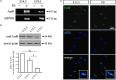
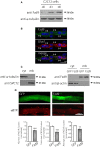
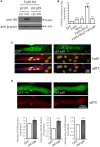
Cumulative evidence indicates that Wnt pathways play crucial and diverse roles to assemble the neuromuscular junction (NMJ), a peripheral synapse characterized by the clustering of acetylcholine receptors (AChR) on postsynaptic densities. The molecular determinants of Wnt effects at the NMJ are still to be fully elucidated. We report here that the Wnt receptor Frizzled-9 (Fzd9) is expressed in developing skeletal muscles during NMJ synaptogenesis. In cultured myotubes, gain- and loss-of-function experiments revealed that Fzd9-mediated signaling impairs the AChR-clustering activity of agrin, an organizer of postsynaptic differentiation. Overexpression of Fzd9 induced the cytosolic accumulation of β-catenin, a key regulator of Wnt signaling. Consistently, Fzd9 and β-catenin localize in the postsynaptic domain of embryonic NMJs in vivo. Our findings represent the first evidence pointing to a crucial role of a Fzd-mediated, β-catenin-dependent signaling on the assembly of the vertebrate NMJ.
Keywords: Frizzled receptors; Wnt proteins; acetylcholine receptor; neuromuscular junction; postsynaptic; skeletal muscle.
Figures
Similar articles
-
Site-directed MT1-MMP trafficking and surface insertion regulate AChR clustering and remodeling at developing NMJs.Elife. 2020 Mar 24;9:e54379. doi: 10.7554/eLife.54379. Elife. 2020. PMID: 32208136 Free PMC article.
-
Wnt/beta-catenin signaling suppresses Rapsyn expression and inhibits acetylcholine receptor clustering at the neuromuscular junction.J Biol Chem. 2008 Aug 1;283(31):21668-75. doi: 10.1074/jbc.M709939200. Epub 2008 Jun 9. J Biol Chem. 2008. PMID: 18541538
-
Tyrosine phosphatases such as SHP-2 act in a balance with Src-family kinases in stabilization of postsynaptic clusters of acetylcholine receptors.BMC Neurosci. 2007 Jul 2;8:46. doi: 10.1186/1471-2202-8-46. BMC Neurosci. 2007. PMID: 17605785 Free PMC article.
-
Crosstalk between Agrin and Wnt signaling pathways in development of vertebrate neuromuscular junction.Dev Neurobiol. 2014 Aug;74(8):828-38. doi: 10.1002/dneu.22190. Epub 2014 Jun 4. Dev Neurobiol. 2014. PMID: 24838312 Review.
-
Structural mechanisms of the agrin-LRP4-MuSK signaling pathway in neuromuscular junction differentiation.Cell Mol Life Sci. 2013 Sep;70(17):3077-88. doi: 10.1007/s00018-012-1209-9. Epub 2012 Nov 22. Cell Mol Life Sci. 2013. PMID: 23178848 Free PMC article. Review.
Cited by
-
Terminal Schwann Cell Aging: Implications for Age-Associated Neuromuscular Dysfunction.Aging Dis. 2021 Apr 1;12(2):494-514. doi: 10.14336/AD.2020.0708. eCollection 2021 Apr. Aging Dis. 2021. PMID: 33815879 Free PMC article. Review.
-
MuSK frizzled-like domain is critical for mammalian neuromuscular junction formation and maintenance.J Neurosci. 2015 Mar 25;35(12):4926-41. doi: 10.1523/JNEUROSCI.3381-14.2015. J Neurosci. 2015. PMID: 25810523 Free PMC article.
-
Synergic action of MicroRNAs and Wnts delivered by motor neuron EVs in promoting AChR clustering.Cell Commun Signal. 2025 Aug 1;23(1):360. doi: 10.1186/s12964-025-02312-x. Cell Commun Signal. 2025. PMID: 40750900 Free PMC article.
-
Frizzled-9+ Supporting Cells Are Progenitors for the Generation of Hair Cells in the Postnatal Mouse Cochlea.Front Mol Neurosci. 2019 Jul 31;12:184. doi: 10.3389/fnmol.2019.00184. eCollection 2019. Front Mol Neurosci. 2019. PMID: 31427926 Free PMC article.
-
Crosstalk among canonical Wnt and Hippo pathway members in skeletal muscle and at the neuromuscular junction.Neural Regen Res. 2025 Sep 1;20(9):2464-2479. doi: 10.4103/NRR.NRR-D-24-00417. Epub 2024 Sep 6. Neural Regen Res. 2025. PMID: 39248171 Free PMC article.
References
-
- Fu A. K., Ip F. C., Fu W. Y., Cheung J., Wang J. H., Yung W. H., et al. (2005). Aberrant motor axon projection, acetylcholine receptor clustering, and neurotransmission in cyclin-dependent kinase 5 null mice. Proc. Natl. Acad. Sci. U.S.A. 102, 15224–15229 10.1073/pnas.0507678102 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

