Wheat germ cell-free system-based production of hemagglutinin-neuraminidase glycoprotein of human parainfluenza virus type 3 for generation and characterization of monoclonal antibody
- PMID: 24860558
- PMCID: PMC4026691
- DOI: 10.3389/fmicb.2014.00208
Wheat germ cell-free system-based production of hemagglutinin-neuraminidase glycoprotein of human parainfluenza virus type 3 for generation and characterization of monoclonal antibody
Abstract
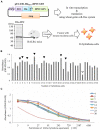
Human parainfluenza virus 3 (HPIV3) commonly causes respiratory disorders in infants and young children. Monoclonal antibodies (MAbs) have been produced to several components of HPIV3 and commercially available. However, the utility of these antibodies for several immunological and proteomic assays for understanding the nature of HPIV3 infection remain to be characterized. Herein, we report the development and characterization of MAbs against hemagglutinin-neuraminidase (HN) of HPIV3. A recombinant full-length HPIV3-HN was successfully synthesized using the wheat-germ cell-free protein production system. After immunization and cell fusion, 36 mouse hybridomas producing MAbs to HPIV3-HN were established. The MAbs obtained were fully characterized using ELISA, immunoblotting, and immunofluorescent analyses. Of the MAbs tested, single clone was found to be applicable in both flow cytometry and immunoprecipitation procedures. By utilizing the antibody, we identified HPIV3-HN binding host proteins via immunoprecipitation-based mass spectrometry analysis. The newly-developed MAbs could thus be a valuable tool for the study of HPIV3 infection as well as the several diagnostic tests of this virus.
Keywords: cell-free protein synthesis; human parainfluenza virus 3; monoclonal antibody; proteomics.
Figures

Similar articles
-
Inhibiting Human Parainfluenza Virus Infection by Preactivating the Cell Entry Mechanism.mBio. 2019 Feb 19;10(1):e02900-18. doi: 10.1128/mBio.02900-18. mBio. 2019. PMID: 30782664 Free PMC article.
-
Bovine parainfluenza virus type 3 (BPIV3) fusion and hemagglutinin-neuraminidase glycoproteins make an important contribution to the restricted replication of BPIV3 in primates.J Virol. 2000 Oct;74(19):8922-9. doi: 10.1128/jvi.74.19.8922-8929.2000. J Virol. 2000. PMID: 10982335 Free PMC article.
-
Evaluation of the replication and immunogenicity of recombinant human parainfluenza virus type 3 vectors expressing up to three foreign glycoproteins.Virology. 2002 May 25;297(1):136-52. doi: 10.1006/viro.2002.1415. Virology. 2002. PMID: 12083844
-
Down-regulation of paramyxovirus hemagglutinin-neuraminidase glycoprotein surface expression by a mutant fusion protein containing a retention signal for the endoplasmic reticulum.J Virol. 1996 Aug;70(8):5005-15. doi: 10.1128/JVI.70.8.5005-5015.1996. J Virol. 1996. PMID: 8764007 Free PMC article.
-
New antiviral approaches for human parainfluenza: Inhibiting the haemagglutinin-neuraminidase.Antiviral Res. 2019 Jul;167:89-97. doi: 10.1016/j.antiviral.2019.04.001. Epub 2019 Apr 3. Antiviral Res. 2019. PMID: 30951732 Review.
Cited by
-
Development of Monoclonal Antibody and Diagnostic Test for Middle East Respiratory Syndrome Coronavirus Using Cell-Free Synthesized Nucleocapsid Antigen.Front Microbiol. 2016 Apr 20;7:509. doi: 10.3389/fmicb.2016.00509. eCollection 2016. Front Microbiol. 2016. PMID: 27148198 Free PMC article.
-
Development of a Monoclonal Antibody Targeting HTLV-1 Envelope gp46 Glycoprotein and Its Application to Near-Infrared Photoimmuno-Antimicrobial Strategy.Viruses. 2022 Sep 29;14(10):2153. doi: 10.3390/v14102153. Viruses. 2022. PMID: 36298708 Free PMC article.
-
Cell-Free Protein Synthesis: A Promising Option for Future Drug Development.BioDrugs. 2020 Jun;34(3):327-348. doi: 10.1007/s40259-020-00417-y. BioDrugs. 2020. PMID: 32198631 Free PMC article. Review.
-
Generation and Utilization of a Monoclonal Antibody against Hepatitis B Virus Core Protein for a Comprehensive Interactome Analysis.Microorganisms. 2022 Nov 30;10(12):2381. doi: 10.3390/microorganisms10122381. Microorganisms. 2022. PMID: 36557634 Free PMC article.
-
Easy Synthesis of Complex Biomolecular Assemblies: Wheat Germ Cell-Free Protein Expression in Structural Biology.Front Mol Biosci. 2021 Mar 25;8:639587. doi: 10.3389/fmolb.2021.639587. eCollection 2021. Front Mol Biosci. 2021. PMID: 33842544 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources

