Rare deleterious mutations of the gene EFR3A in autism spectrum disorders
- PMID: 24860643
- PMCID: PMC4032628
- DOI: 10.1186/2040-2392-5-31
Rare deleterious mutations of the gene EFR3A in autism spectrum disorders
Abstract
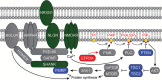
Background: Whole-exome sequencing studies in autism spectrum disorder (ASD) have identified de novo mutations in novel candidate genes, including the synaptic gene Eighty-five Requiring 3A (EFR3A). EFR3A is a critical component of a protein complex required for the synthesis of the phosphoinositide PtdIns4P, which has a variety of functions at the neural synapse. We hypothesized that deleterious mutations in EFR3A would be significantly associated with ASD.
Methods: We conducted a large case/control association study by deep resequencing and analysis of whole-exome data for coding and splice site variants in EFR3A. We determined the potential impact of these variants on protein structure and function by a variety of conservation measures and analysis of the Saccharomyces cerevisiae Efr3 crystal structure. We also analyzed the expression pattern of EFR3A in human brain tissue.
Results: Rare nonsynonymous mutations in EFR3A were more common among cases (16 / 2,196 = 0.73%) than matched controls (12 / 3,389 = 0.35%) and were statistically more common at conserved nucleotides based on an experiment-wide significance threshold (P = 0.0077, permutation test). Crystal structure analysis revealed that mutations likely to be deleterious were also statistically more common in cases than controls (P = 0.017, Fisher exact test). Furthermore, EFR3A is expressed in cortical neurons, including pyramidal neurons, during human fetal brain development in a pattern consistent with ASD-related genes, and it is strongly co-expressed (P < 2.2 × 10(-16), Wilcoxon test) with a module of genes significantly associated with ASD.
Conclusions: Rare deleterious mutations in EFR3A were found to be associated with ASD using an experiment-wide significance threshold. Synaptic phosphoinositide metabolism has been strongly implicated in syndromic forms of ASD. These data for EFR3A strengthen the evidence for the involvement of this pathway in idiopathic autism.
Keywords: Autism spectrum disorder; EFR3A; Genetics; Phosphoinositide metabolism; Rare variants; Synapse.
Figures


References
-
- American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders. 5. Washington, DC: American Psychiatric Association; 2013. Autism Spectrum Disorder; pp. 50–59.
-
- Autism and Developmental Disabilities Monitoring Network Surveillance Year 2008 Principal Investigators and Centers for Disease Control and Prevention. Prevalence of autism spectrum disorders – autism and developmental disabilities monitoring network, 14 sites, United States. MMWR Surveill Summ. 2008;2012(61):1–19. - PubMed
-
- Sanders SJ, Murtha MT, Gupta AR, Murdoch JD, Raubeson MJ, Willsey AJ, Ercan-Sencicek AG, DiLullo NM, Parikshak NN, Stein JL, Walker MF, Ober GT, Teran NA, Song Y, El-Fishawy P, Murtha R, Choi M, Overton JD, Bjornson RD, Carrierio NJ, Meyer KA, Bilguvar K, Mane SM, Sestan N, Lifton RP, Gunel M, Roeder K, Geschwind DH, Devlin B, State MW. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485:237–241. doi: 10.1038/nature10945. - DOI - PMC - PubMed
-
- Iossifov I, Ronemus M, Levy D, Wang Z, Hakker I, Rosenbaum J, Yamrom B, Lee YH, Narzisi G, Leotta A, Kendall J, Grabowska E, Ma B, Marks S, Rodgers L, Stepansky A, Troge J, Andrews P, Bekritsky M, Pradhan K, Ghiban E, Kramer M, Parla J, Demeter R, Fulton LL, Fulton RS, Magrini VJ, Ye K, Darnell JC, Darnell RB. et al.De novo gene disruptions in children on the autistic spectrum. Neuron. 2012;74:285–299. doi: 10.1016/j.neuron.2012.04.009. - DOI - PMC - PubMed
-
- Neale BM, Kou Y, Liu L, Ma’ayan A, Samocha KE, Sabo A, Lin CF, Stevens C, Wang LS, Makarov V, Polak P, Yoon S, Maguire J, Crawford EL, Campbell NG, Geller ET, Valladares O, Schafer C, Liu H, Zhao T, Cai G, Lihm J, Dannenfelser O, Jabado Z, Peralta U, Nagaswamy U, Muzny D, Reid JG, Newsham I, Wu Y. et al.Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485:242–245. doi: 10.1038/nature11011. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

