Differential (14)N/(15)N-Labeling of Peptides Using N-Terminal Charge Derivatization with a High-Proton Affinity for Straightforward de novo Peptide Sequencing
- PMID: 24860714
- PMCID: PMC3967017
- DOI: 10.5702/massspectrometry.A0024
Differential (14)N/(15)N-Labeling of Peptides Using N-Terminal Charge Derivatization with a High-Proton Affinity for Straightforward de novo Peptide Sequencing
Abstract
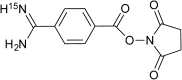
While de novo peptide sequencing is essential in many situations, it remains a difficult task. This is because peptide fragmentation results in complicated and often incomplete product ion spectra. In a previous study, we demonstrated that N-terminal charge derivatization with 4-amidinobenzoic acid (Aba) resulted in improved peptide fragmentation under low-energy CID conditions. However, even with this derivatization, some ambiguity exists, due to difficulties in discriminating between N- and C-terminal fragments. In this study, to specifically identify b-ions from complex product ion spectra, the differential (14)N/(15)N-labeling of peptides was performed using Aba derivatization. (15)N-Labeled Aba was synthesized in the form of a succinimide ester. Peptides were derivatized individually with (14)N-Aba or (15)N-Aba and analyzed by ESI-MS/MS using a linear ion trap-Orbitrap hybrid FTMS system. The N-terminal fragments (i.e., b-ions) were then identified based on m/z differences arising from isotope labeling. By comparing the spectra between (14)N- and (15)N-Aba derivatized peptides, b-ions could be successfully identified based on the m/z shifts, which provided reliable sequencing results for all of the peptides examined in this study. The method developed in this study allows the easy and reliable de novo sequencing of peptides, which is useful in peptidomics and proteomics studies.
Keywords: MS/MS; de novo sequencing; derivatization; fragmentation; isotope labeling.
Figures




Similar articles
-
Improving peptide fragmentation by N-terminal derivatization with high proton affinity.Rapid Commun Mass Spectrom. 2011 May 15;25(9):1130-40. doi: 10.1002/rcm.4962. Rapid Commun Mass Spectrom. 2011. PMID: 21488112
-
N-Terminal Derivatization with Structures Having High Proton Affinity for Discrimination between Leu and Ile Residues in Peptides by High-Energy Collision-Induced Dissociation.Mass Spectrom (Tokyo). 2016;5(1):A0051. doi: 10.5702/massspectrometry.A0051. Epub 2016 Nov 25. Mass Spectrom (Tokyo). 2016. PMID: 27900234 Free PMC article.
-
N-Terminal chemical derivatization of peptides with 4-formyl-benzenesulfonic acid and electrospray positive and negative tandem mass spectrometry with high abundance of b-ion series.Rapid Commun Mass Spectrom. 2023 Jul 30;37(14):e9534. doi: 10.1002/rcm.9534. Rapid Commun Mass Spectrom. 2023. PMID: 37147273
-
De novo sequencing of peptides by MS/MS.Proteomics. 2010 Feb;10(4):634-49. doi: 10.1002/pmic.200900459. Proteomics. 2010. PMID: 19953542 Review.
-
Trends in the Design of New Isobaric Labeling Reagents for Quantitative Proteomics.Molecules. 2019 Feb 15;24(4):701. doi: 10.3390/molecules24040701. Molecules. 2019. PMID: 30781343 Free PMC article. Review.
References
-
- 1) V. H. Wysocki, K. A. Resing, Q. Zhang, G. Cheng. Mass spectrometry of peptides and proteins. Methods 35: 211–222, 2005 - PubMed
-
- 2) A. I. Nesvizhskii. Protein identification by tandem mass spectrometry and sequence database searching. Methods Mol. Biol. 367: 87–119, 2007 - PubMed
-
- 3) J. Seidler, N. Zinn, M. E. Boehm, W. D. Lehmann. De novo sequencing of peptides by MS/MS. Proteomics 10: 634–649, 2010 - PubMed
-
- 4) G. Menschaert, T. T. M. Vandekerckhove, G. Baggerman, L. Schoofs, W. Luyten, W. Van Criekinge. Peptidomics coming of age: A review of contributions from a bioinformatics angle. J. Proteome Res. 9: 2051–2061, 2010 - PubMed
-
- 5) K. D. Roth, Z. H. Huang, N. Sadagopan, J. T. Watson. Charge derivatization of peptides for analysis by mass spectrometry. Mass Spectrom. Rev. 17: 255–274, 1998 - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

