Lentiviral protein transduction with genome-modifying HIV-1 integrase-I-PpoI fusion proteins: studies on specificity and cytotoxicity
- PMID: 24860818
- PMCID: PMC4016911
- DOI: 10.1155/2014/379340
Lentiviral protein transduction with genome-modifying HIV-1 integrase-I-PpoI fusion proteins: studies on specificity and cytotoxicity
Abstract
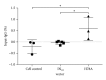
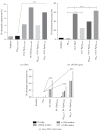
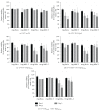
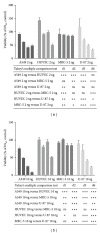
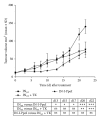
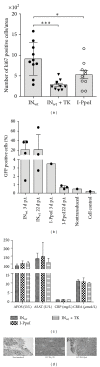
Rare-cutting endonucleases, such as the I-PpoI, can be used for the induction of double strand breaks (DSBs) in genome editing and targeted integration based on homologous recombination. For therapeutic approaches, the specificity and the pattern of off-target effects are of high importance in these techniques. For its applications, the endonuclease needs to be transported into the target cell nucleus, where the mechanism of transport may affect its function. Here, we have studied the lentiviral protein transduction of the integrase (IN)-PpoI fusion protein using the cis-packaging method. In genome-wide interaction studies, IN-fusion proteins were verified to bind their target sequence containing 28S ribosomal RNA (rRNA) genes with a 100-fold enrichment, despite the well-documented behavior of IN to be tethered into various genomic areas by host-cell factors. In addition, to estimate the applicability of the method, DSB-induced cytotoxic effects with different vector endonuclease configurations were studied in a panel of cells. Varying the amount and activity of endonuclease enabled the adjustment of ratio between the induced DSBs and transported DNA. In cell studies, certain cancerous cell lines were especially prone to DSBs in rRNA genes, which led us to test the protein transduction in a tumour environment in an in vivo study. In summary, the results highlight the potential of lentiviral vectors (LVVs) for the nuclear delivery of endonucleases.
Figures
Similar articles
-
Characterization of a new IN-I-PpoI fusion protein and a homology-arm containing transgene cassette that improve transgene expression persistence and 28S rRNA gene-targeted insertion of lentiviral vectors.PLoS One. 2023 Jan 20;18(1):e0280894. doi: 10.1371/journal.pone.0280894. eCollection 2023. PLoS One. 2023. PMID: 36662822 Free PMC article.
-
Development of Lentiviral Vectors for Targeted Integration and Protein Delivery.Methods Mol Biol. 2016;1448:185-98. doi: 10.1007/978-1-4939-3753-0_14. Methods Mol Biol. 2016. PMID: 27317182
-
rDNA-directed integration by an HIV-1 integrase--I-PpoI fusion protein.Nucleic Acids Res. 2013 Mar 1;41(5):e61. doi: 10.1093/nar/gks1438. Epub 2012 Dec 28. Nucleic Acids Res. 2013. PMID: 23275537 Free PMC article.
-
Non-integrating lentiviral vectors.Curr Gene Ther. 2008 Dec;8(6):430-7. doi: 10.2174/156652308786848012. Curr Gene Ther. 2008. PMID: 19075626 Review.
-
Integration-deficient lentiviral vectors: a slow coming of age.Mol Ther. 2009 Aug;17(8):1316-32. doi: 10.1038/mt.2009.122. Epub 2009 Jun 2. Mol Ther. 2009. PMID: 19491821 Free PMC article. Review.
Cited by
-
Characterization of a new IN-I-PpoI fusion protein and a homology-arm containing transgene cassette that improve transgene expression persistence and 28S rRNA gene-targeted insertion of lentiviral vectors.PLoS One. 2023 Jan 20;18(1):e0280894. doi: 10.1371/journal.pone.0280894. eCollection 2023. PLoS One. 2023. PMID: 36662822 Free PMC article.
-
Time-Restricted PiggyBac DNA Transposition by Transposase Protein Delivery Using Lentivirus-Derived Nanoparticles.Mol Ther Nucleic Acids. 2018 Jun 1;11:253-262. doi: 10.1016/j.omtn.2018.02.006. Epub 2018 Mar 30. Mol Ther Nucleic Acids. 2018. PMID: 29858060 Free PMC article.
-
Efficient Nuclease-Directed Integration of Lentivirus Vectors into the Human Ribosomal DNA Locus.Mol Ther. 2020 Aug 5;28(8):1858-1875. doi: 10.1016/j.ymthe.2020.05.019. Epub 2020 May 23. Mol Ther. 2020. PMID: 32504545 Free PMC article.
References
-
- Ford KG, Souberbielle BE, Darling D, Farzaneh F. Protein transduction: an alternative to genetic intervention? Gene Therapy. 2001;8(1):1–4. - PubMed
-
- Ingvarsdottir K, Blaho JA. Association of the herpes simplex virus major tegument structural protein VP22 with chromatin. Biochimica et Biophysica Acta. 2010;1799(3-4):200–206. - PubMed
-
- Noguchi H, Matsumoto S. Protein transduction technology: a novel therapeutic perspective. Acta Medica Okayama. 2006;60(1):1–11. - PubMed
-
- Wurm FM. Production of recombinant protein therapeutics in cultivated mammalian cells. Nature Biotechnology. 2004;22(11):1393–1398. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

