ProBiS-ligands: a web server for prediction of ligands by examination of protein binding sites
- PMID: 24861616
- PMCID: PMC4086080
- DOI: 10.1093/nar/gku460
ProBiS-ligands: a web server for prediction of ligands by examination of protein binding sites
Abstract
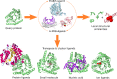
The ProBiS-ligands web server predicts binding of ligands to a protein structure. Starting with a protein structure or binding site, ProBiS-ligands first identifies template proteins in the Protein Data Bank that share similar binding sites. Based on the superimpositions of the query protein and the similar binding sites found, the server then transposes the ligand structures from those sites to the query protein. Such ligand prediction supports many activities, e.g. drug repurposing. The ProBiS-ligands web server, an extension of the ProBiS web server, is open and free to all users at http://probis.cmm.ki.si/ligands.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Yuriev E., Ramsland P.A. Latest developments in molecular docking: 2010–2011 in review. J. Mol. Recognit. 2013;26:215–239. - PubMed
-
- Haupt V.J., Schroeder M. Old friends in new guise: repositioning of known drugs with structural bioinformatics. Briefings Bioinform. 2011;12:312–326. - PubMed
-
- Moriaud F., Richard S.B., Adcock S.A., Chanas-Martin L., Surgand J.-S., Jelloul M.B., Delfaud F. Identify drug repurposing candidates by mining the Protein Data Bank. Briefings Bioinform. 2011;12:336–340. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

