Quantitative telomerase enzyme activity determination using droplet digital PCR with single cell resolution
- PMID: 24861623
- PMCID: PMC4117742
- DOI: 10.1093/nar/gku439
Quantitative telomerase enzyme activity determination using droplet digital PCR with single cell resolution
Abstract
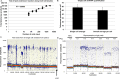
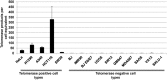
The telomere repeat amplification protocol (TRAP) for the human reverse transcriptase, telomerase, is a PCR-based assay developed two decades ago and is still used for routine determination of telomerase activity. The TRAP assay can only reproducibly detect ∼ 2-fold differences and is only quantitative when compared to internal standards and reference cell lines. The method generally involves laborious radioactive gel electrophoresis and is not conducive to high-throughput analyzes. Recently droplet digital PCR (ddPCR) technologies have become available that allow for absolute quantification of input deoxyribonucleic acid molecules following PCR. We describe the reproducibility and provide several examples of a droplet digital TRAP (ddTRAP) assay for telomerase activity, including quantitation of telomerase activity in single cells, telomerase activity across several common telomerase positive cancer cells lines and in human primary peripheral blood mononuclear cells following mitogen stimulation. Adaptation of the TRAP assay to digital format allows accurate and reproducible quantification of the number of telomerase-extended products (i.e. telomerase activity; 57.8 ± 7.5) in a single HeLa cell. The tools developed in this study allow changes in telomerase enzyme activity to be monitored on a single cell basis and may have utility in designing novel therapeutic approaches that target telomerase.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures






Similar articles
-
ddTRAP: A Method for Sensitive and Precise Quantification of Telomerase Activity.Methods Mol Biol. 2018;1768:513-529. doi: 10.1007/978-1-4939-7778-9_29. Methods Mol Biol. 2018. PMID: 29717462 Free PMC article.
-
Droplet Digital TRAP (ddTRAP): Adaptation of the Telomere Repeat Amplification Protocol to Droplet Digital Polymerase Chain Reaction.J Vis Exp. 2019 May 3;(147). doi: 10.3791/59550. J Vis Exp. 2019. PMID: 31107456
-
Assessment and quantification of telomerase enzyme activity.Methods Mol Biol. 2013;965:215-31. doi: 10.1007/978-1-62703-239-1_14. Methods Mol Biol. 2013. PMID: 23296661
-
Detection of telomerase activity by the TRAP assay and its variants and alternatives.Clin Chim Acta. 2006 Sep;371(1-2):25-31. doi: 10.1016/j.cca.2006.02.039. Epub 2006 Apr 17. Clin Chim Acta. 2006. PMID: 16616059 Review.
-
Real-time detection and quantification of telomerase activity utilizing energy transfer primers.Methods Mol Biol. 2006;335:157-69. doi: 10.1385/1-59745-069-3:157. Methods Mol Biol. 2006. PMID: 16785627 Review.
Cited by
-
Analysis of TERT Isoforms across TCGA, GTEx and CCLE Datasets.Cancers (Basel). 2021 Apr 13;13(8):1853. doi: 10.3390/cancers13081853. Cancers (Basel). 2021. PMID: 33924498 Free PMC article.
-
Preliminary development of an assay for detection of TERT expression, telomere length, and telomere elongation in single cells.PLoS One. 2018 Dec 5;13(12):e0206525. doi: 10.1371/journal.pone.0206525. eCollection 2018. PLoS One. 2018. PMID: 30517099 Free PMC article.
-
A highly sensitive and specific method for the screening detection of genetically modified organisms based on digital PCR without pretreatment.Sci Rep. 2015 Aug 4;5:12715. doi: 10.1038/srep12715. Sci Rep. 2015. PMID: 26239916 Free PMC article.
-
Intronic Cis-Element DR8 in hTERT Is Bound by Splicing Factor SF3B4 and Regulates hTERT Splicing in Non-Small Cell Lung Cancer.Mol Cancer Res. 2022 Oct 4;20(10):1574-1588. doi: 10.1158/1541-7786.MCR-21-0058. Mol Cancer Res. 2022. PMID: 35852380 Free PMC article.
-
DNA damage response at telomeres boosts the transcription of SARS-CoV-2 receptor ACE2 during aging.EMBO Rep. 2022 Feb 3;23(2):e53658. doi: 10.15252/embr.202153658. Epub 2021 Dec 2. EMBO Rep. 2022. PMID: 34854526 Free PMC article.
References
-
- Kim N.W., Piatyszek M.A., Prowse K.R., Harley C.B., West M.D., Ho P.L., Coviello G.M., Wright W.E., Weinrich S.L., Shay J.W. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. - PubMed
-
- Harley C.B., Kim N.W., Prowse K.R., Weinrich S.L., Hirsch K.S., West M.D., Bacchetti S., Hirte H.W., Counter C.M., Greider C.W., et al. Telomerase, cell immortality, and cancer. Cold Spring Harb. Symp. Quant. Biol. 1994;59:307–315. - PubMed
-
- Greider C.W. Mammalian telomere dynamics: healing, fragmentation shortening and stabilization. Curr. Opin. Genet. Dev. 1994;4:203–211. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

