Epigenetics and nutritional environmental signals
- PMID: 24861811
- PMCID: PMC4072902
- DOI: 10.1093/icb/icu049
Epigenetics and nutritional environmental signals
Abstract
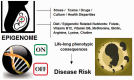
All terrestrial life is influenced by multi-directional flows of information about its environment, enabling malleable phenotypic change through signals, chemical processes, or various forms of energy that facilitate acclimatization. Billions of biological co-inhabitants of the earth, including all plants and animals, collectively make up a genetic/epigenetic ecosystem by which adaptation/survival (inputs and outputs) are highly interdependent on one another. As an ecosystem, the solar system, rotation of the planets, changes in sunlight, and gravitational pull influence cyclic epigenetic transitions and chromatin remodeling that constitute biological circadian rhythms controlling senescence. In humans, adverse environmental conditions such as poverty, stress, alcohol, malnutrition, exposure to pollutants generated from industrialization, man-made chemicals, and use of synthetic drugs can lead to maladaptive epigenetic-related illnesses with disease-specific genes being atypically activated or silenced. Nutrition and dietary practices are one of the largest facets in epigenetic-related metabolism, where specific "epi-nutrients" can stabilize the genome, given established roles in DNA methylation, histone modification, and chromatin remodeling. Moreover, food-based "epi-bioactive" constituents may reverse maladaptive epigenetic patterns, not only prior to conception and during fetal/early postnatal development but also through adulthood. In summary, in contrast to a static genomic DNA structure, epigenetic changes are potentially reversible, raising the hope for therapeutic and/or dietary interventions that can reverse deleterious epigenetic programing as a means to prevent or treat major illnesses.
Published by Oxford University Press on behalf of the Society for Integrative and Comparative Biology 2014. This work is written by US Government employees and is in the public domain in the US.
Figures
Similar articles
-
Epigenetics in an ecotoxicological context.Mutat Res Genet Toxicol Environ Mutagen. 2014 Apr;764-765:36-45. doi: 10.1016/j.mrgentox.2013.08.008. Epub 2013 Sep 1. Mutat Res Genet Toxicol Environ Mutagen. 2014. PMID: 24004878 Review.
-
The Impact of Nutrition and Environmental Epigenetics on Human Health and Disease.Int J Mol Sci. 2018 Nov 1;19(11):3425. doi: 10.3390/ijms19113425. Int J Mol Sci. 2018. PMID: 30388784 Free PMC article. Review.
-
Epigenetic variation and environmental change.J Exp Bot. 2015 Jun;66(12):3541-8. doi: 10.1093/jxb/eru502. Epub 2015 Feb 17. J Exp Bot. 2015. PMID: 25694547 Review.
-
Epigenetic modifications of gene expression by lifestyle and environment.Arch Pharm Res. 2017 Nov;40(11):1219-1237. doi: 10.1007/s12272-017-0973-3. Epub 2017 Oct 17. Arch Pharm Res. 2017. PMID: 29043603 Review.
-
Transgenerational inheritance: how impacts to the epigenetic and genetic information of parents affect offspring health.Hum Reprod Update. 2019 Sep 11;25(5):518-540. doi: 10.1093/humupd/dmz017. Hum Reprod Update. 2019. PMID: 31374565
Cited by
-
Maternal Malnutrition and Elevated Disease Risk in Offspring.Nutrients. 2024 Aug 8;16(16):2614. doi: 10.3390/nu16162614. Nutrients. 2024. PMID: 39203750 Free PMC article. Review.
-
Cardiometabolic Effects of Postnatal High-Fat Diet Consumption in Offspring Exposed to Maternal Protein Restriction In Utero.Front Physiol. 2022 May 10;13:829920. doi: 10.3389/fphys.2022.829920. eCollection 2022. Front Physiol. 2022. PMID: 35620602 Free PMC article. Review.
-
Epigenetic Mechanisms of the Aging Human Retina.J Exp Neurosci. 2016 Feb 3;9(Suppl 2):51-79. doi: 10.4137/JEN.S25513. eCollection 2015. J Exp Neurosci. 2016. PMID: 26966390 Free PMC article. Review.
-
Maternal vitamin D deficiency during pregnancy results in insulin resistance in rat offspring, which is associated with inflammation and Iκbα methylation.Diabetologia. 2014 Oct;57(10):2165-72. doi: 10.1007/s00125-014-3316-7. Epub 2014 Jul 2. Diabetologia. 2014. PMID: 24985146
-
A Susceptible Period of Photic Day-Night Rhythm Loss in Common Marmoset Social Behavior Development.Front Behav Neurosci. 2021 Feb 2;14:539411. doi: 10.3389/fnbeh.2020.539411. eCollection 2020. Front Behav Neurosci. 2021. PMID: 33603653 Free PMC article.
References
-
- Altmann S, Murani E, Schwerin M, Metges CC, Wimmers K, Ponsuksili S. Maternal dietary protein restriction and excess affects offspring gene expression and methylation of non-SMC subunits of condensin I in liver and skeletal muscle. Epigenetics. 2012;7:239–52. - PubMed
-
- Andersen SO, Halberstadt ML, Borgford-Parnell N. Stratospheric ozone, global warming, and the principle of unintended consequences–an ongoing science and policy success story. J Air Waste Manag Assoc. 2013;63:607–47. - PubMed
-
- Barnes SK, Ozanne SE. Pathways linking the early environment to long-term health and lifespan. Progr Biophysics Molecular Biol. 2011;106:323–36. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

