The first 1000 cultured species of the human gastrointestinal microbiota
- PMID: 24861948
- PMCID: PMC4262072
- DOI: 10.1111/1574-6976.12075
The first 1000 cultured species of the human gastrointestinal microbiota
Abstract
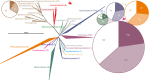
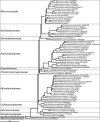
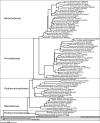
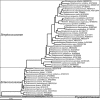
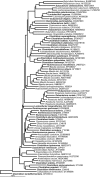
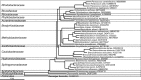
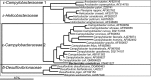
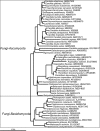
The microorganisms that inhabit the human gastrointestinal tract comprise a complex ecosystem with functions that significantly contribute to our systemic metabolism and have an impact on health and disease. In line with its importance, the human gastrointestinal microbiota has been extensively studied. Despite the fact that a significant part of the intestinal microorganisms has not yet been cultured, presently over 1000 different microbial species that can reside in the human gastrointestinal tract have been identified. This review provides a systematic overview and detailed references of the total of 1057 intestinal species of Eukarya (92), Archaea (8) and Bacteria (957), based on the phylogenetic framework of their small subunit ribosomal RNA gene sequences. Moreover, it unifies knowledge about the prevalence, abundance, stability, physiology, genetics and the association with human health of these gastrointestinal microorganisms, which is currently scattered over a vast amount of literature published in the last 150 years. This detailed physiological and genetic information is expected to be instrumental in advancing our knowledge of the gastrointestinal microbiota. Moreover, it opens avenues for future comparative and functional metagenomic and other high-throughput approaches that need a systematic and physiological basis to have an impact.
Keywords: diversity; function; gastrointestinal; gut; microbiome; microbiota.
© 2014 The Authors. FEMS Microbiology Reviews published by John Wiley & Sons Ltd on behalf of Federation of European Microbiological Societies.
Figures




















References
-
- Alexieff A. Sur la nature des formations dites ‘Kystes de Trichomonas intestinalis’. CR Soc Biol. 1911;71:296–298.
-
- Allison MJ, Dawson KA, Mayberry WR. Foss JG. Oxalobacter formigenes gen. nov., sp. nov.: oxalate-degrading anaerobes that inhabit the gastrointestinal tract. Arch Microbiol. 1985;141:1–7. - PubMed
-
- Anderson HW. Yeast-like fungi of the human intestinal tract. J Infect Dis. 1917;21:341–386.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

