Insights from cerebellar transcriptomic analysis into the pathogenesis of ataxia
- PMID: 24862029
- PMCID: PMC4469030
- DOI: 10.1001/jamaneurol.2014.756
Insights from cerebellar transcriptomic analysis into the pathogenesis of ataxia
Abstract
Importance: The core clinical and neuropathological feature of the autosomal dominant spinocerebellar ataxias (SCAs) is cerebellar degeneration. Mutations in the known genes explain only 50% to 60% of SCA cases. To date, no effective treatments exist, and the knowledge of drug-treatable molecular pathways is limited. The examination of overlapping mechanisms and the interpretation of how ataxia genes interact will be important in the discovery of potential disease-modifying agents.
Objectives: To address the possible relationships among known SCA genes, predict their functions, identify overlapping pathways, and provide a framework for candidate gene discovery using whole-transcriptome expression data.
Design, setting, and participants: We have used a systems biology approach based on whole-transcriptome gene expression analysis. As part of the United Kingdom Brain Expression Consortium, we analyzed the expression profile of 788 brain samples obtained from 101 neuropathologically healthy individuals (10 distinct brain regions each). Weighted gene coexpression network analysis was used to cluster 24 SCA genes into gene coexpression modules in an unsupervised manner. The overrepresentation of SCA transcripts in modules identified in the cerebellum was assessed. Enrichment analysis was performed to infer the functions and molecular pathways of genes in biologically relevant modules.
Main outcomes and measures: Molecular functions and mechanisms implicating SCA genes, as well as lists of relevant coexpressed genes as potential candidates for novel SCA causative or modifier genes.
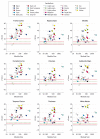
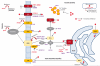
Results: Two cerebellar gene coexpression modules were statistically enriched in SCA transcripts (P = .021 for the tan module and P = 2.87 × 10-5 for the light yellow module) and contained established granule and Purkinje cell markers, respectively. One module includes genes involved in the ubiquitin-proteasome system and contains SCA genes usually associated with a complex phenotype, while the other module encloses many genes important for calcium homeostasis and signaling and contains SCA genes associated mostly with pure ataxia.
Conclusions and relevance: Using normal gene expression in the human brain, we identified significant cell types and pathways in SCA pathogenesis. The overrepresentation of genes involved in calcium homeostasis and signaling may indicate an important target for therapy in the future. Furthermore, the gene networks provide new candidate genes for ataxias or novel genes that may be critical for cerebellar function.
Figures

Comment in
-
Coexpression networks predict ataxia genes.JAMA Neurol. 2014 Jul 1;71(7):825-8. doi: 10.1001/jamaneurol.2014.757. JAMA Neurol. 2014. PMID: 24861589 No abstract available.
References
-
- Schöls L, Bauer P, Schmidt T, Schulte T, Riess O. Autosomal dominant cerebellar ataxias: clinical features, genetics, and pathogenesis. Lancet Neurol. 2004;3(5):291–304. - PubMed
-
- Dueñas AM, Goold R, Giunti P. Molecular pathogenesis of spinocerebellar ataxias. Brain. 2006;129(pt 6):1357–1370. - PubMed
-
- Soong BW, Paulson HL. Spinocerebellar ataxias: an update. Curr Opin Neurol. 2007;20(4):438–446. - PubMed
-
- Durr A. Autosomal dominant cerebellar ataxias: polyglutamine expansions and beyond. Lancet Neurol. 2010;9(9):885–894. - PubMed
-
- Wang JL, Yang X, Xia K, et al. TGM6 identified as a novel causative gene of spinocerebellar ataxias using exome sequencing. Brain. 2010;133(pt 12):3510–3518. - PubMed
Publication types
MeSH terms
Grants and funding
- G0901254/MRC_/Medical Research Council/United Kingdom
- MR/K01417X/1/MRC_/Medical Research Council/United Kingdom
- MR/K000608/1/MRC_/Medical Research Council/United Kingdom
- G1001253/MRC_/Medical Research Council/United Kingdom
- 089698/WT_/Wellcome Trust/United Kingdom
- MR/J004758/1/MRC_/Medical Research Council/United Kingdom
- G0601440/MRC_/Medical Research Council/United Kingdom
- G0802462/MRC_/Medical Research Council/United Kingdom
- G108/638/MRC_/Medical Research Council/United Kingdom
- MC_G1000735/MRC_/Medical Research Council/United Kingdom
- G0802760/MRC_/Medical Research Council/United Kingdom
- WT089698/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

