Organellophagy: eliminating cellular building blocks via selective autophagy
- PMID: 24862571
- PMCID: PMC4033777
- DOI: 10.1083/jcb.201402054
Organellophagy: eliminating cellular building blocks via selective autophagy
Abstract
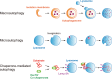
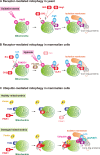
Maintenance of organellar quality and quantity is critical for cellular homeostasis and adaptation to variable environments. Emerging evidence demonstrates that this kind of control is achieved by selective elimination of organelles via autophagy, termed organellophagy. Organellophagy consists of three key steps: induction, cargo tagging, and sequestration, which involve signaling pathways, organellar landmark molecules, and core autophagy-related proteins, respectively. In addition, posttranslational modifications such as phosphorylation and ubiquitination play important roles in recruiting and tailoring the autophagy machinery to each organelle. The basic principles underlying organellophagy are conserved from yeast to mammals, highlighting its biological relevance in eukaryotic cells.
© 2014 Okamoto.
Figures

References
-
- Birgisdottir A.B., Lamark T., Johansen T. 2013. The LIR motif - crucial for selective autophagy. J. Cell Sci. 126:3237–3247 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

