A TOGL domain specifically targets yeast CLASP to kinetochores to stabilize kinetochore microtubules
- PMID: 24862575
- PMCID: PMC4033772
- DOI: 10.1083/jcb.201310018
A TOGL domain specifically targets yeast CLASP to kinetochores to stabilize kinetochore microtubules
Abstract
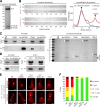
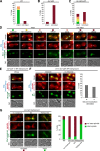
Cytoplasmic linker-associated proteins (CLASPs) are proposed to function in cell division based on their ability to bind tubulin via arrayed tumor overexpressed gene (TOG)-like (TOGL) domains. Structure predictions suggest that CLASPs have at least two TOGL domains. We show that only TOGL2 of Saccharomyces cerevisiae CLASP Stu1 binds to tubulin and is required for polymerization of spindle microtubules (MTs) in vivo. In contrast, TOGL1 recruits Stu1 to kinetochores (KTs), where it is essential for the stability and tension-dependent regulation of KT MTs. Stu1 is also recruited to spindle MTs by different mechanisms depending on the mitotic phase: in metaphase, Stu1 binds directly to the MT lattice, whereas in anaphase, it is localized indirectly to the spindle midzone. In both phases, the activity of TOGL2 is essential for interpolar MT stability, whereas TOGL1 is not involved. Thus, the two TOGL domains of yeast CLASP have different activities and execute distinct mitotic functions.
© 2014 Funk et al.
Figures







References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

