In vitro reconstitution of a cellular phase-transition process that involves the mRNA decapping machinery
- PMID: 24862735
- PMCID: PMC4320757
- DOI: 10.1002/anie.201402885
In vitro reconstitution of a cellular phase-transition process that involves the mRNA decapping machinery
Abstract
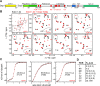
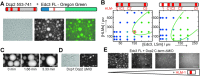
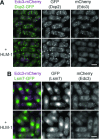
In eukaryotic cells, components of the 5' to 3' mRNA degradation machinery can undergo a rapid phase transition. The resulting cytoplasmic foci are referred to as processing bodies (P-bodies). The molecular details of the self-aggregation process are, however, largely undetermined. Herein, we use a bottom-up approach that combines NMR spectroscopy, isothermal titration calorimetry, X-ray crystallography, and fluorescence microscopy to probe if mRNA degradation factors can undergo phase transitions in vitro. We show that the Schizosaccharomyces pombe Dcp2 mRNA decapping enzyme, its prime activator Dcp1, and the scaffolding proteins Edc3 and Pdc1 are sufficient to reconstitute a phase-separation process. Intermolecular interactions between the Edc3 LSm domain and at least 10 helical leucine-rich motifs in Dcp2 and Pdc1 build the core of the interaction network. We show that blocking of these interactions interferes with the clustering behavior, both in vitro and in vivo.
Keywords: NMR spectroscopy; phase transitions; processing bodies; protein-protein interactions; self-assembly.
© 2014 The Authors. Published by Wiley-VCH Verlag GmbH & Co. KGaA. This is an open access article under the terms of the Creative Commons Attribution Non-Commercial NoDerivs License, which permits use and distribution in any medium, provided the original work is properly cited, the use is non-commercial and no modifications or adaptations are made.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

