Regulation of mammalian siderophore 2,5-DHBA in the innate immune response to infection
- PMID: 24863067
- PMCID: PMC4042634
- DOI: 10.1084/jem.20132629
Regulation of mammalian siderophore 2,5-DHBA in the innate immune response to infection
Abstract
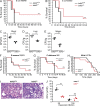
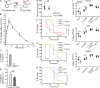
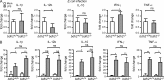
Competition for iron influences host-pathogen interactions. Pathogens secrete small iron-binding moieties, siderophores, to acquire host iron. In response, the host secretes siderophore-binding proteins, such as lipocalin 24p3, which limit siderophore-mediated iron import into bacteria. Mammals produce 2,5-dihydroxy benzoic acid, a compound that resembles a bacterial siderophore. Our data suggest that bacteria use both mammalian and bacterial siderophores. In support of this idea, supplementation with mammalian siderophore enhances bacterial growth in vitro. In addition, mice lacking the mammalian siderophore resist E. coli infection. Finally, we show that the host responds to infection by suppressing siderophore synthesis while up-regulating lipocalin 24p3 expression via TLR signaling. Thus, reciprocal regulation of 24p3 and mammalian siderophore is a protective mechanism limiting microbial access to iron.
© 2014 Liu et al.
Figures




Comment in
-
Microbial hijacking of mammalian iron shuttling.J Exp Med. 2014 Jun 2;211(6):1009. doi: 10.1084/jem.2116insight3. J Exp Med. 2014. PMID: 24890116 Free PMC article. No abstract available.
References
-
- Albrecht-Gary A.M., Crumbliss A.L. 1998. Coordination chemistry of siderophores: thermodynamics and kinetics of iron chelation and release. Met. Ions Biol. Syst. 35:239–327 - PubMed
-
- Aung H.T., Schroder K., Himes S.R., Brion K., van Zuylen W., Trieu A., Suzuki H., Hayashizaki Y., Hume D.A., Sweet M.J., Ravasi T. 2006. LPS regulates proinflammatory gene expression in macrophages by altering histone deacetylase expression. FASEB J. 20:1315–1327 10.1096/fj.05-5360com - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

