WNT5A enhances resistance of melanoma cells to targeted BRAF inhibitors
- PMID: 24865425
- PMCID: PMC4071371
- DOI: 10.1172/JCI70156
WNT5A enhances resistance of melanoma cells to targeted BRAF inhibitors
Abstract
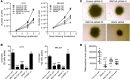
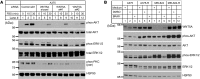
About half of all melanomas harbor a mutation that results in a constitutively active BRAF kinase mutant (BRAF(V600E/K)) that can be selectively inhibited by targeted BRAF inhibitors (BRAFis). While patients treated with BRAFis initially exhibit measurable clinical improvement, the majority of patients eventually develop drug resistance and relapse. Here, we observed marked elevation of WNT5A in a subset of tumors from patients exhibiting disease progression on BRAFi therapy. WNT5A transcript and protein were also elevated in BRAFi-resistant melanoma cell lines generated by long-term in vitro treatment with BRAFi. RNAi-mediated reduction of endogenous WNT5A in melanoma decreased cell growth, increased apoptosis in response to BRAFi challenge, and decreased the activity of prosurvival AKT signaling. Conversely, overexpression of WNT5A promoted melanoma growth, tumorigenesis, and activation of AKT signaling. Similarly to WNT5A knockdown, knockdown of the WNT receptors FZD7 and RYK inhibited growth, sensitized melanoma cells to BRAFi, and reduced AKT activation. Together, these findings suggest that chronic BRAF inhibition elevates WNT5A expression, which promotes AKT signaling through FZD7 and RYK, leading to increased growth and therapeutic resistance. Furthermore, increased WNT5A expression in BRAFi-resistant melanomas correlates with a specific transcriptional signature, which identifies potential therapeutic targets to reduce clinical BRAFi resistance.
Figures





References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

