Virus-encoded microRNAs facilitate gammaherpesvirus latency and pathogenesis in vivo
- PMID: 24865551
- PMCID: PMC4045068
- DOI: 10.1128/mBio.00981-14
Virus-encoded microRNAs facilitate gammaherpesvirus latency and pathogenesis in vivo
Abstract
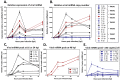
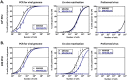
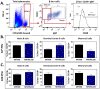
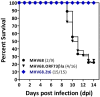
Gammaherpesviruses, including Epstein-Barr virus (EBV), Kaposi sarcoma-associated herpesvirus (KSHV, or HHV-8), and murine gammaherpesvirus 68 (MHV68, γHV68, or MuHV-4), are B cell-tropic pathogens that each encode at least 12 microRNAs (miRNAs). It is predicted that these regulatory RNAs facilitate infection by suppressing host target genes involved in a wide range of key cellular pathways. However, the precise contribution that gammaherpesvirus miRNAs make to viral life cycle and pathogenesis in vivo is unknown. MHV68 infection of mice provides a highly useful system to dissect the function of specific viral elements in the context of both asymptomatic infection and disease. Here, we report (i) analysis of in vitro and in vivo MHV68 miRNA expression, (ii) generation of an MHV68 miRNA mutant with reduced expression of all 14 pre-miRNA stem-loops, and (iii) comprehensive phenotypic characterization of the miRNA mutant virus in vivo. The profile of MHV68 miRNAs detected in infected cell lines varied with cell type and did not fully recapitulate the profile from cells latently infected in vivo. The miRNA mutant virus, MHV68.Zt6, underwent normal lytic replication in vitro and in vivo, demonstrating that the MHV68 miRNAs are dispensable for acute replication. During chronic infection, MHV68.Zt6 was attenuated for latency establishment, including a specific defect in memory B cells. Finally, MHV68.Zt6 displayed a striking attenuation in the development of lethal pneumonia in mice deficient in IFN-γ. These data indicate that the MHV68 miRNAs may facilitate virus-driven maturation of infected B cells and implicate the miRNAs as a critical determinant of gammaherpesvirus-associated disease.
Importance: Gammaherpesviruses such as EBV and KSHV are widespread pathogens that establish lifelong infections and are associated with the development of numerous types of diseases, including cancer. Gammaherpesviruses encode many small noncoding RNAs called microRNAs (miRNAs). It is predicted that gammaherpesvirus miRNAs facilitate infection and disease by suppressing host target transcripts involved in a wide range of key cellular pathways; however, the precise contribution that these regulatory RNAs make to in vivo virus infection and pathogenesis is unknown. Here, we generated a mutated form of murine gammaherpesvirus (MHV68) to dissect the function of gammaherpesvirus miRNAs in vivo. We demonstrate that the MHV68 miRNAs were dispensable for short-term virus replication but were important for establishment of lifelong infection in the key virus reservoir of memory B cells. Moreover, the MHV68 miRNAs were essential for the development of virus-associated pneumonia, implicating them as a critical component of gammaherpesvirus-associated disease.
Copyright © 2014 Feldman et al.
Figures





Similar articles
-
Identification of murine gammaherpesvirus 68 miRNA-mRNA hybrids reveals miRNA target conservation among gammaherpesviruses including host translation and protein modification machinery.PLoS Pathog. 2019 Aug 8;15(8):e1007843. doi: 10.1371/journal.ppat.1007843. eCollection 2019 Aug. PLoS Pathog. 2019. PMID: 31393953 Free PMC article.
-
The Antagonism between the Murine Gammaherpesvirus Protein Kinase and Global Interferon Regulatory Factor 1 Expression Shapes the Establishment of Chronic Infection.J Virol. 2022 Oct 26;96(20):e0126022. doi: 10.1128/jvi.01260-22. Epub 2022 Sep 28. J Virol. 2022. PMID: 36169331 Free PMC article.
-
Lytic Replication and Reactivation from B Cells Is Not Required for Establishing or Maintaining Gammaherpesvirus Latency In Vivo.J Virol. 2022 Jun 22;96(12):e0069022. doi: 10.1128/jvi.00690-22. Epub 2022 Jun 1. J Virol. 2022. PMID: 35647668 Free PMC article.
-
Murine Gammaherpesvirus 68: A Small Animal Model for Gammaherpesvirus-Associated Diseases.Adv Exp Med Biol. 2017;1018:225-236. doi: 10.1007/978-981-10-5765-6_14. Adv Exp Med Biol. 2017. PMID: 29052141 Review.
-
Gamma interferon blocks gammaherpesvirus reactivation from latency in a cell type-specific manner.J Virol. 2007 Jun;81(11):6134-40. doi: 10.1128/JVI.00108-07. Epub 2007 Mar 14. J Virol. 2007. PMID: 17360749 Free PMC article. Review.
Cited by
-
The Role of microRNAs in the Pathogenesis of Herpesvirus Infection.Viruses. 2016 Jun 2;8(6):156. doi: 10.3390/v8060156. Viruses. 2016. PMID: 27271654 Free PMC article. Review.
-
The Structure-To-Function Relationships of Gammaherpesvirus-Encoded Long Non-Coding RNAs and Their Contributions to Viral Pathogenesis.Noncoding RNA. 2018 Sep 26;4(4):24. doi: 10.3390/ncrna4040024. Noncoding RNA. 2018. PMID: 30261651 Free PMC article. Review.
-
Viruses and miRNAs: More Friends than Foes.Front Microbiol. 2017 May 15;8:824. doi: 10.3389/fmicb.2017.00824. eCollection 2017. Front Microbiol. 2017. PMID: 28555130 Free PMC article. Review.
-
KSHV-Mediated Angiogenesis in Tumor Progression.Viruses. 2016 Jul 20;8(7):198. doi: 10.3390/v8070198. Viruses. 2016. PMID: 27447661 Free PMC article. Review.
-
Connivance, Complicity, or Collusion? The Role of Noncoding RNAs in Promoting Gammaherpesvirus Tumorigenesis.Trends Cancer. 2018 Nov;4(11):729-740. doi: 10.1016/j.trecan.2018.09.005. Epub 2018 Oct 10. Trends Cancer. 2018. PMID: 30352676 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

