IraL is an RssB anti-adaptor that stabilizes RpoS during logarithmic phase growth in Escherichia coli and Shigella
- PMID: 24865554
- PMCID: PMC4045071
- DOI: 10.1128/mBio.01043-14
IraL is an RssB anti-adaptor that stabilizes RpoS during logarithmic phase growth in Escherichia coli and Shigella
Abstract
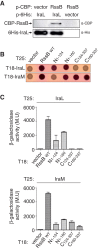
RpoS (σ(S)), the general stress response sigma factor, directs the expression of genes under a variety of stressful conditions. Control of the cellular σ(S) concentration is critical for appropriately scaled σ(S)-dependent gene expression. One way to maintain appropriate levels of σ(S) is to regulate its stability. Indeed, σ(S) degradation is catalyzed by the ClpXP protease and the recognition of σ(S) by ClpXP depends on the adaptor protein RssB. Three anti-adaptors (IraD, IraM, and IraP) exist in Escherichia coli K-12; each interacts with RssB and inhibits RssB activity under different stress conditions, thereby stabilizing σ(S). Unlike K-12, some E. coli isolates, including uropathogenic E. coli strain CFT073, show comparable cellular levels of σ(S) during the logarithmic and stationary growth phases, suggesting that there are differences in the regulation of σ(S) levels among E. coli strains. Here, we describe IraL, an RssB anti-adaptor that stabilizes σ(S) during logarithmic phase growth in CFT073 and other E. coli and Shigella strains. By immunoblot analyses, we show that IraL affects the levels and stability of σ(S) during logarithmic phase growth. By computational and PCR-based analyses, we reveal that iraL is found in many E. coli pathotypes but not in laboratory-adapted strains. Finally, by bacterial two-hybrid and copurification analyses, we demonstrate that IraL interacts with RssB by a mechanism distinct from that used by other characterized anti-adaptors. We introduce a fourth RssB anti-adaptor found in E. coli species and suggest that differences in the regulation of σ(S) levels may contribute to host and niche specificity in pathogenic and nonpathogenic E. coli strains.
Importance: Bacteria must cope with a variety of environmental conditions in order to survive. RpoS (σ(S)), the general stress response sigma factor, directs the expression of many genes under stressful conditions in both pathogenic and nonpathogenic Escherichia coli strains. The regulation of σ(S) levels and activity allows appropriately scaled σ(S)-dependent gene expression. Here, we describe IraL, an RssB anti-adaptor that, unlike previously described anti-adaptors, stabilizes σ(S) during the logarithmic growth phase in the absence of additional stress. We also demonstrate that iraL is found in a large number of E. coli and Shigella isolates. These data suggest that strains containing iraL are able to initiate σ(S)-dependent gene expression under conditions under which strains without iraL cannot. Therefore, IraL-mediated σ(S) stabilization may contribute to host and niche specificity in E. coli.
Copyright © 2014 Hryckowian et al.
Figures


Similar articles
-
Modulating RssB activity: IraP, a novel regulator of sigma(S) stability in Escherichia coli.Genes Dev. 2006 Apr 1;20(7):884-97. doi: 10.1101/gad.1400306. Genes Dev. 2006. PMID: 16600914 Free PMC article.
-
Anti-adaptors use distinct modes of binding to inhibit the RssB-dependent turnover of RpoS (σ(S)) by ClpXP.Front Mol Biosci. 2015 Apr 23;2:15. doi: 10.3389/fmolb.2015.00015. eCollection 2015. Front Mol Biosci. 2015. PMID: 25988182 Free PMC article.
-
Regulation of RpoS proteolysis in Escherichia coli: the response regulator RssB is a recognition factor that interacts with the turnover element in RpoS.Proc Natl Acad Sci U S A. 1999 May 25;96(11):6439-44. doi: 10.1073/pnas.96.11.6439. Proc Natl Acad Sci U S A. 1999. PMID: 10339606 Free PMC article.
-
Back to log phase: sigma S as a global regulator in the osmotic control of gene expression in Escherichia coli.Mol Microbiol. 1996 Sep;21(5):887-93. doi: 10.1046/j.1365-2958.1996.511405.x. Mol Microbiol. 1996. PMID: 8885260 Review.
-
Signal transduction and regulatory mechanisms involved in control of the sigma(S) (RpoS) subunit of RNA polymerase.Microbiol Mol Biol Rev. 2002 Sep;66(3):373-95, table of contents. doi: 10.1128/MMBR.66.3.373-395.2002. Microbiol Mol Biol Rev. 2002. PMID: 12208995 Free PMC article. Review.
Cited by
-
The General Stress Response σS Is Regulated by a Partner Switch in the Gram-negative Bacterium Shewanella oneidensis.J Biol Chem. 2016 Dec 9;291(50):26151-26163. doi: 10.1074/jbc.M116.751933. Epub 2016 Nov 3. J Biol Chem. 2016. PMID: 27810894 Free PMC article.
-
dsdA Does Not Affect Colonization of the Murine Urinary Tract by Escherichia coli CFT073.PLoS One. 2015 Sep 14;10(9):e0138121. doi: 10.1371/journal.pone.0138121. eCollection 2015. PLoS One. 2015. PMID: 26366567 Free PMC article.
-
RpoS and the bacterial general stress response.Microbiol Mol Biol Rev. 2024 Mar 27;88(1):e0015122. doi: 10.1128/mmbr.00151-22. Epub 2024 Feb 27. Microbiol Mol Biol Rev. 2024. PMID: 38411096 Free PMC article. Review.
-
Trouble is coming: Signaling pathways that regulate general stress responses in bacteria.J Biol Chem. 2019 Aug 2;294(31):11685-11700. doi: 10.1074/jbc.REV119.005593. Epub 2019 Jun 13. J Biol Chem. 2019. PMID: 31197038 Free PMC article. Review.
-
Regulated Proteolysis in Vibrio cholerae Allowing Rapid Adaptation to Stress Conditions.Front Cell Infect Microbiol. 2019 Jun 21;9:214. doi: 10.3389/fcimb.2019.00214. eCollection 2019. Front Cell Infect Microbiol. 2019. PMID: 31293982 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

