HuR regulates alternative splicing of the TRA2β gene in human colon cancer cells under oxidative stress
- PMID: 24865968
- PMCID: PMC4135568
- DOI: 10.1128/MCB.00333-14
HuR regulates alternative splicing of the TRA2β gene in human colon cancer cells under oxidative stress
Abstract
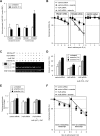
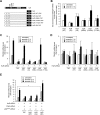
Hu antigen R (HuR) regulates stress responses through stabilizing and/or facilitating the translation of target mRNAs. The human TRA2β gene encodes splicing factor transformer 2β (Tra2β) and generates 5 mRNA isoforms (TRA2β1 to -5) through alternative splicing. Exposure of HCT116 colon cancer cells to sodium arsenite stimulated checkpoint kinase 2 (Chk2)- and mitogen-activated protein kinase p38 (p38(MAPK))-mediated phosphorylation of HuR at positions S88 and T118. This induced an association between HuR and the 39-nucleotide (nt) proximal region of TRA2β exon 2, generating a TRA2β4 mRNA that includes exon 2, which has multiple premature stop codons. HuR knockdown or Chk2/p38(MAPK) double knockdown inhibited the arsenite-stimulated production of TRA2β4 and increased Tra2β protein, facilitating Tra2β-dependent inclusion of exons in target pre-mRNAs. The effects of HuR knockdown or Chk2/p38(MAPK) double knockdown were also confirmed using a TRA2β minigene spanning exons 1 to 4, and the effects disappeared when the 39-nt region was deleted from the minigene. In endogenous HuR knockdown cells, the overexpression of a HuR mutant that could not be phosphorylated (with changes of serine to alanine at position 88 [S88A], S100A, and T118A) blocked the associated TRA2β4 interaction and TRA2β4 generation, while the overexpression of a phosphomimetic HuR (with mutations S88D, S100D, and T118D) restored the TRA2β4-related activities. Our findings revealed the potential role of nuclear HuR in the regulation of alternative splicing programs under oxidative stress.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures






Similar articles
-
Oxidative stress-induced alternative splicing of transformer 2beta (SFRS10) and CD44 pre-mRNAs in gastric epithelial cells.Am J Physiol Cell Physiol. 2009 Aug;297(2):C330-8. doi: 10.1152/ajpcell.00009.2009. Epub 2009 May 13. Am J Physiol Cell Physiol. 2009. PMID: 19439532
-
Ets1 and heat shock factor 1 regulate transcription of the Transformer 2β gene in human colon cancer cells.J Gastroenterol. 2013 Nov;48(11):1222-33. doi: 10.1007/s00535-012-0745-2. Epub 2013 Jan 30. J Gastroenterol. 2013. PMID: 23361474
-
HnRNPA1 interacts with G-quadruplex in the TRA2B promoter and stimulates its transcription in human colon cancer cells.Sci Rep. 2019 Jul 16;9(1):10276. doi: 10.1038/s41598-019-46659-x. Sci Rep. 2019. PMID: 31311954 Free PMC article.
-
How does Tra2β protein regulate tissue-specific RNA splicing?Biochem Soc Trans. 2012 Aug;40(4):784-8. doi: 10.1042/BST20120036. Biochem Soc Trans. 2012. PMID: 22817734 Free PMC article. Review.
-
Regulation of the mRNA-binding protein HuR by posttranslational modification: spotlight on phosphorylation.Curr Protein Pept Sci. 2012 Jun;13(4):380-90. doi: 10.2174/138920312801619439. Curr Protein Pept Sci. 2012. PMID: 22708484 Review.
Cited by
-
Transite: A Computational Motif-Based Analysis Platform That Identifies RNA-Binding Proteins Modulating Changes in Gene Expression.Cell Rep. 2020 Aug 25;32(8):108064. doi: 10.1016/j.celrep.2020.108064. Cell Rep. 2020. PMID: 32846122 Free PMC article.
-
Nucleolin facilitates nuclear retention of an ultraconserved region containing TRA2β4 and accelerates colon cancer cell growth.Oncotarget. 2018 Jun 1;9(42):26817-26833. doi: 10.18632/oncotarget.25510. eCollection 2018 Jun 1. Oncotarget. 2018. PMID: 29928487 Free PMC article.
-
Ultraconserved region-containing Transformer 2β4 controls senescence of colon cancer cells.Oncogenesis. 2016 Apr 4;5(4):e213. doi: 10.1038/oncsis.2016.18. Oncogenesis. 2016. PMID: 27043659 Free PMC article.
-
Functional Network Analysis Reveals the Relevance of SKIIP in the Regulation of Alternative Splicing by p38 SAPK.Cell Rep. 2019 Apr 16;27(3):847-859.e6. doi: 10.1016/j.celrep.2019.03.060. Cell Rep. 2019. PMID: 30995481 Free PMC article.
-
CircNUP54 promotes hepatocellular carcinoma progression via facilitating HuR cytoplasmic export and stabilizing BIRC3 mRNA.Cell Death Dis. 2024 Mar 5;15(3):191. doi: 10.1038/s41419-024-06570-4. Cell Death Dis. 2024. PMID: 38443362 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
