Histidine methylation of yeast ribosomal protein Rpl3p is required for proper 60S subunit assembly
- PMID: 24865971
- PMCID: PMC4135575
- DOI: 10.1128/MCB.01634-13
Histidine methylation of yeast ribosomal protein Rpl3p is required for proper 60S subunit assembly
Abstract
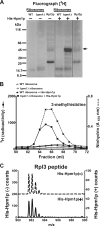
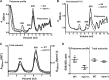
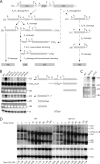
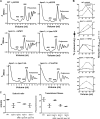
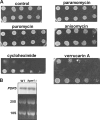
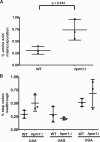
Histidine protein methylation is an unusual posttranslational modification. In the yeast Saccharomyces cerevisiae, the large ribosomal subunit protein Rpl3p is methylated at histidine 243, a residue that contacts the 25S rRNA near the P site. Rpl3p methylation is dependent upon the presence of Hpm1p, a candidate seven-beta-strand methyltransferase. In this study, we elucidated the biological activities of Hpm1p in vitro and in vivo. Amino acid analyses reveal that Hpm1p is responsible for all of the detectable protein histidine methylation in yeast. The modification is found on a polypeptide corresponding to the size of Rpl3p in ribosomes and in a nucleus-containing organelle fraction but was not detected in proteins of the ribosome-free cytosol fraction. In vitro assays demonstrate that Hpm1p has methyltransferase activity on ribosome-associated but not free Rpl3p, suggesting that its activity depends on interactions with ribosomal components. hpm1 null cells are defective in early rRNA processing, resulting in a deficiency of 60S subunits and translation initiation defects that are exacerbated in minimal medium. Cells lacking Hpm1p are resistant to cycloheximide and verrucarin A and have decreased translational fidelity. We propose that Hpm1p plays a role in the orchestration of the early assembly of the large ribosomal subunit and in faithful protein production.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures

References
-
- Lapeyre B. 2005. Conserved ribosomal RNA modification and their putative roles in ribosome biogenesis and translation, p 263–284 In Grosjean H. (ed), Fine-tuning of RNA functions by modification and editing. Springer, Berlin, Germany
-
- Johansson MO, Byström A. 2005. Transfer RNA modifications and modifying enzymes in Saccharomyces cerevisiae, p 87–120 In Grosjean H. (ed), Fine-tuning of RNA functions by modification and editing. Springer, Berlin, Germany
-
- Bokar J. 2005. The biosynthesis and functional roles of methylated nucleosides in eukaryotic mRNA, p 141–177 In Grosjean H. (ed), Fine-tuning of RNA functions by modification and editing. Springer, Berlin, Germany
-
- Katz JE, Dlakic M, Clarke SG. 2003. Automated identification of putative methyltransferases from genomic open reading frames. Mol. Cell. Proteomics 2:525–540 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
