Sequestration of multiple RNA recognition motif-containing proteins by C9orf72 repeat expansions
- PMID: 24866055
- PMCID: PMC4065024
- DOI: 10.1093/brain/awu120
Sequestration of multiple RNA recognition motif-containing proteins by C9orf72 repeat expansions
Abstract
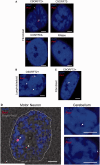
GGGGCC repeat expansions of C9orf72 represent the most common genetic variant of amyotrophic lateral sclerosis and frontotemporal degeneration, but the mechanism of pathogenesis is unclear. Recent reports have suggested that the transcribed repeat might form toxic RNA foci that sequester various RNA processing proteins. Consensus as to the identity of the binding partners is missing and whole neuronal proteome investigation is needed. Using RNA fluorescence in situ hybridization we first identified nuclear and cytoplasmic RNA foci in peripheral and central nervous system biosamples from patients with amyotrophic lateral sclerosis with a repeat expansion of C9orf72 (C9orf72+), but not from those patients without a repeat expansion of C9orf72 (C9orf72-) or control subjects. Moreover, in the cases examined, the distribution of foci-positive neurons correlated with the clinical phenotype (t-test P < 0.05). As expected, RNA foci are ablated by RNase treatment. Interestingly, we identified foci in fibroblasts from an asymptomatic C9orf72+ carrier. We next performed pulldown assays, with GGGGCC5, in conjunction with mass spectrometry analysis, to identify candidate binding partners of the GGGGCC repeat expansion. Proteins containing RNA recognition motifs and involved in splicing, messenger RNA nuclear export and/or translation were significantly enriched. Immunohistochemistry in central nervous system tissue from C9orf72+ patients with amyotrophic lateral sclerosis demonstrated co-localization of RNA foci with SRSF2, hnRNP H1/F, ALYREF and hnRNP A1 in cerebellar granule cells and with SRSF2, hnRNP H1/F and ALYREF in motor neurons, the primary target of pathology in amyotrophic lateral sclerosis. Direct binding of proteins to GGGGCC repeat RNA was confirmed in vitro by ultraviolet-crosslinking assays. Co-localization was only detected in a small proportion of RNA foci, suggesting dynamic sequestration rather than irreversible binding. Additional immunohistochemistry demonstrated that neurons with and without RNA foci were equally likely to show nuclear depletion of TDP-43 (χ(2) P = 0.75) or poly-GA dipeptide repeat protein inclusions (χ(2) P = 0.46). Our findings suggest two non-exclusive pathogenic mechanisms: (i) functional depletion of RNA-processing proteins resulting in disruption of messenger RNA splicing; and (ii) licensing of expanded C9orf72 pre-messenger RNA for nuclear export by inappropriate association with messenger RNA export adaptor protein(s) leading to cytoplasmic repeat associated non-ATG translation and formation of potentially toxic dipeptide repeat protein.
Keywords: amyotrophic lateral sclerosis; fluorescence imaging; genetics; pathology.
© The Author (2014). Published by Oxford University Press on behalf of the Guarantors of Brain.
Figures






References
-
- Bengoechea R, Tapia O, Casafont I, Berciano J, Lafarga M, Berciano MT. Nuclear speckles are involved in nuclear aggregation of PABPN1 and in the pathophysiology of oculopharyngeal muscular dystrophy. Neurobiol Dis. 2012;46:118–29. - PubMed
-
- Byrne S, Heverin M, Elamin M, Walsh C, Hardiman O. Intermediate repeat expansion length in C9orf72 may be pathological in amyotrophic lateral sclerosis. Amyotroph Lateral Scler Frontotemporal Degener. 2013;15:148–50. - PubMed
-
- Casafont I, Bengoechea R, Tapia O, Berciano MT, Lafarga M. TDP-43 localizes in mRNA transcription and processing sites in mammalian neurons. J Struct Biol. 2009;167:235–41. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- MR/L016451/1/MRC_/Medical Research Council/United Kingdom
- G1100540/MRC_/Medical Research Council/United Kingdom
- G0502157/MRC_/Medical Research Council/United Kingdom
- MR/K003771/1/MRC_/Medical Research Council/United Kingdom
- G0400074/MRC_/Medical Research Council/United Kingdom
- COOPER-KNOCK/FEB12/942-795/MNDA_/Motor Neurone Disease Association/United Kingdom
- MR/K000039/1/MRC_/Medical Research Council/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- BB/D011795/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- G0900652/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

