Rapid mapping of interactions between Human SNX-BAR proteins measured in vitro by AlphaScreen and single-molecule spectroscopy
- PMID: 24866125
- PMCID: PMC4159646
- DOI: 10.1074/mcp.M113.037275
Rapid mapping of interactions between Human SNX-BAR proteins measured in vitro by AlphaScreen and single-molecule spectroscopy
Abstract
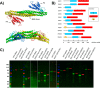
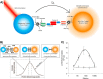
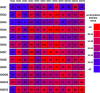
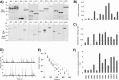
Protein dimerization and oligomerization is commonly used by nature to increase the structural and functional complexity of proteins. Regulated protein assembly is essential to transfer information in signaling, transcriptional, and membrane trafficking events. Here we show that a combination of cell-free protein expression, a proximity based interaction assay (AlphaScreen), and single-molecule fluorescence allow rapid mapping of homo- and hetero-oligomerization of proteins. We have applied this approach to the family of BAR domain-containing sorting nexin (SNX-BAR) proteins, which are essential regulators of membrane trafficking and remodeling in all eukaryotes. Dimerization of BAR domains is essential for creating a concave structure capable of sensing and inducing membrane curvature. We have systematically mapped 144 pairwise interactions between the human SNX-BAR proteins and generated an interaction matrix of preferred dimerization partners for each family member. We find that while nine SNX-BAR proteins are able to form homo-dimers, several including the retromer-associated SNX1, SNX2, and SNX5 require heteromeric interactions for dimerization. SNX2, SNX4, SNX6, and SNX8 show a promiscuous ability to bind other SNX-BAR proteins and we also observe a novel interaction with the SNX3 protein which lacks the BAR domain structure.
© 2014 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures
Similar articles
-
SNX-BAR-mediated endosome tubulation is co-ordinated with endosome maturation.Traffic. 2012 Jan;13(1):94-107. doi: 10.1111/j.1600-0854.2011.01297.x. Epub 2011 Oct 31. Traffic. 2012. PMID: 21973056
-
Cargo-selective SNX-BAR proteins mediate retromer trimer independent retrograde transport.J Cell Biol. 2017 Nov 6;216(11):3677-3693. doi: 10.1083/jcb.201702137. Epub 2017 Sep 21. J Cell Biol. 2017. PMID: 28935632 Free PMC article.
-
Molecular basis for SNX-BAR-mediated assembly of distinct endosomal sorting tubules.EMBO J. 2012 Nov 28;31(23):4466-80. doi: 10.1038/emboj.2012.283. Epub 2012 Oct 19. EMBO J. 2012. PMID: 23085988 Free PMC article.
-
The retromer, sorting nexins and the plant endomembrane protein trafficking.J Cell Sci. 2018 Jan 29;131(2):jcs203695. doi: 10.1242/jcs.203695. J Cell Sci. 2018. PMID: 29061884 Review.
-
SNX-PXA-RGS-PXC Subfamily of SNXs in the Regulation of Receptor-Mediated Signaling and Membrane Trafficking.Int J Mol Sci. 2021 Feb 26;22(5):2319. doi: 10.3390/ijms22052319. Int J Mol Sci. 2021. PMID: 33652569 Free PMC article. Review.
Cited by
-
Cell-free formation and interactome analysis of caveolae.J Cell Biol. 2018 Jun 4;217(6):2141-2165. doi: 10.1083/jcb.201707004. Epub 2018 May 1. J Cell Biol. 2018. PMID: 29716956 Free PMC article.
-
Identification of intracellular cavin target proteins reveals cavin-PP1alpha interactions regulate apoptosis.Nat Commun. 2019 Jul 22;10(1):3279. doi: 10.1038/s41467-019-11111-1. Nat Commun. 2019. PMID: 31332168 Free PMC article.
-
Chlamydia exploits filopodial capture and a macropinocytosis-like pathway for host cell entry.PLoS Pathog. 2018 May 4;14(5):e1007051. doi: 10.1371/journal.ppat.1007051. eCollection 2018 May. PLoS Pathog. 2018. PMID: 29727463 Free PMC article.
-
Confocal Spectroscopy to Study Dimerization, Oligomerization and Aggregation of Proteins: A Practical Guide.Int J Mol Sci. 2016 Apr 30;17(5):655. doi: 10.3390/ijms17050655. Int J Mol Sci. 2016. PMID: 27144560 Free PMC article. Review.
-
Functional domain analysis of SOX18 transcription factor using a single-chain variable fragment-based approach.MAbs. 2018 May/Jun;10(4):596-606. doi: 10.1080/19420862.2018.1451288. Epub 2018 Apr 16. MAbs. 2018. PMID: 29648920 Free PMC article.
References
-
- Marianayagam N. J., Sunde M., Matthews J. M. (2004) The power of two: protein dimerization in biology. Trends Biochem. Sci. 29, 618–625 - PubMed
-
- Seet L. F., Hong W. (2006) The Phox (PX) domain proteins and membrane traffic. Biochim. Biophys. Acta 1761, 878–896 - PubMed
-
- Teasdale R. D., Collins B. M. (2012) Insights into the PX (phox-homology) domain and SNX (sorting nexin) protein families: structures, functions, and roles in disease. Biochem. J. 441, 39–59 - PubMed
-
- Cullen P. J. (2011) Phosphoinositides and the regulation of tubular-based endosomal sorting. Biochem. Soc. Trans. 39, 839–850 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

