Natural variation reveals that intracellular distribution of ELF3 protein is associated with function in the circadian clock
- PMID: 24867215
- PMCID: PMC4071560
- DOI: 10.7554/eLife.02206
Natural variation reveals that intracellular distribution of ELF3 protein is associated with function in the circadian clock
Abstract
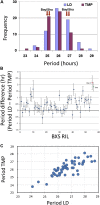
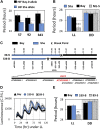
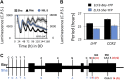
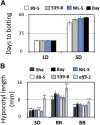
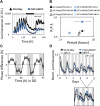
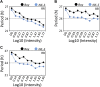
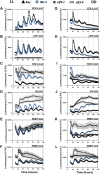
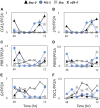
Natural selection of variants within the Arabidopsis thaliana circadian clock can be attributed to adaptation to varying environments. To define a basis for such variation, we examined clock speed in a reporter-modified Bay-0 x Shakdara recombinant inbred line and localized heritable variation. Extensive variation led us to identify EARLY FLOWERING3 (ELF3) as a major quantitative trait locus (QTL). The causal nucleotide polymorphism caused a short-period phenotype under light and severely dampened rhythm generation in darkness, and entrainment alterations resulted. We found that ELF3-Sha protein failed to properly localize to the nucleus, and its ability to accumulate in darkness was compromised. Evidence was provided that the ELF3-Sha allele originated in Central Asia. Collectively, we showed that ELF3 protein plays a vital role in defining its light-repressor action in the circadian clock and that its functional abilities are largely dependent on its cellular localization.
Keywords: QTL mapping/cloning; arabidopsis; cell biology; circadian clock; eQTL; evolutionary biology; genomics; plant biology; population analysis.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

