Muscleblind-like 1 (Mbnl1) regulates pre-mRNA alternative splicing during terminal erythropoiesis
- PMID: 24869935
- PMCID: PMC4110662
- DOI: 10.1182/blood-2013-12-542209
Muscleblind-like 1 (Mbnl1) regulates pre-mRNA alternative splicing during terminal erythropoiesis
Abstract
The scope and roles of regulated isoform gene expression during erythroid terminal development are poorly understood. We identified hundreds of differentiation-associated isoform changes during terminal erythropoiesis. Sequences surrounding cassette exons of skipped exon events are enriched for motifs bound by the Muscleblind-like (MBNL) family of splicing factors. Knockdown of Mbnl1 in cultured murine fetal liver erythroid progenitors resulted in a strong block in erythroid differentiation and disrupted the developmentally regulated exon skipping of Ndel1 mRNA, which is bound by MBNL1 and critical for erythroid terminal proliferation. These findings reveal an unanticipated scope of the alternative splicing program and the importance of Mbnl1 during erythroid terminal differentiation.
© 2014 by The American Society of Hematology.
Figures


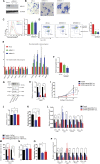
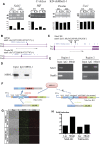

Comment in
-
Erythroid mRNA processing: a "splice of life".Blood. 2014 Jul 24;124(4):474-5. doi: 10.1182/blood-2014-06-579649. Blood. 2014. PMID: 25061168 Free PMC article.
References
-
- Chateauvieux S, Grigorakaki C, Morceau F, Dicato M, Diederich M. Erythropoietin, erythropoiesis and beyond. Biochem Pharmacol. 2011;82(10):1291–1303. - PubMed
-
- Visconte V, Makishima H, Jankowska A, et al. SF3B1, a splicing factor is frequently mutated in refractory anemia with ring sideroblasts. Leukemia. 2012;26(3):542–545. - PubMed
-
- Malcovati L, Papaemmanuil E, Bowen DT, et al. Chronic Myeloid Disorders Working Group of the International Cancer Genome Consortium and of the Associazione Italiana per la Ricerca sul Cancro Gruppo Italiano Malattie Mieloproliferative. Clinical significance of SF3B1 mutations in myelodysplastic syndromes and myelodysplastic/myeloproliferative neoplasms. Blood. 2011;118(24):6239–6246. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

