Accurate design of co-assembling multi-component protein nanomaterials
- PMID: 24870237
- PMCID: PMC4137318
- DOI: 10.1038/nature13404
Accurate design of co-assembling multi-component protein nanomaterials
Abstract
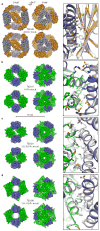
The self-assembly of proteins into highly ordered nanoscale architectures is a hallmark of biological systems. The sophisticated functions of these molecular machines have inspired the development of methods to engineer self-assembling protein nanostructures; however, the design of multi-component protein nanomaterials with high accuracy remains an outstanding challenge. Here we report a computational method for designing protein nanomaterials in which multiple copies of two distinct subunits co-assemble into a specific architecture. We use the method to design five 24-subunit cage-like protein nanomaterials in two distinct symmetric architectures and experimentally demonstrate that their structures are in close agreement with the computational design models. The accuracy of the method and the number and variety of two-component materials that it makes accessible suggest a route to the construction of functional protein nanomaterials tailored to specific applications.
Conflict of interest statement
The authors declare no competing financial interests.
Figures







Similar articles
-
Computational design of self-assembling protein nanomaterials with atomic level accuracy.Science. 2012 Jun 1;336(6085):1171-4. doi: 10.1126/science.1219364. Science. 2012. PMID: 22654060 Free PMC article.
-
Hierarchical design of pseudosymmetric protein nanocages.Nature. 2025 Feb;638(8050):553-561. doi: 10.1038/s41586-024-08360-6. Epub 2024 Dec 18. Nature. 2025. PMID: 39695230 Free PMC article.
-
Accurate design of megadalton-scale two-component icosahedral protein complexes.Science. 2016 Jul 22;353(6297):389-94. doi: 10.1126/science.aaf8818. Science. 2016. PMID: 27463675 Free PMC article.
-
Geometric Principles for Designing Highly Symmetric Self-Assembling Protein Nanomaterials.Annu Rev Biophys. 2017 May 22;46:23-42. doi: 10.1146/annurev-biophys-070816-033928. Epub 2017 Feb 23. Annu Rev Biophys. 2017. PMID: 28301774 Review.
-
Practical approaches to designing novel protein assemblies.Curr Opin Struct Biol. 2013 Aug;23(4):632-8. doi: 10.1016/j.sbi.2013.06.002. Epub 2013 Jul 1. Curr Opin Struct Biol. 2013. PMID: 23827813 Review.
Cited by
-
Protein-protein interaction prediction with deep learning: A comprehensive review.Comput Struct Biotechnol J. 2022 Sep 19;20:5316-5341. doi: 10.1016/j.csbj.2022.08.070. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 36212542 Free PMC article. Review.
-
A roadmap for biocatalysis - functional and spatial orchestration of enzyme cascades.Microb Biotechnol. 2016 Sep;9(5):601-9. doi: 10.1111/1751-7915.12386. Epub 2016 Jul 15. Microb Biotechnol. 2016. PMID: 27418373 Free PMC article. Review.
-
Protein-directed self-assembly of a fullerene crystal.Nat Commun. 2016 Apr 26;7:11429. doi: 10.1038/ncomms11429. Nat Commun. 2016. PMID: 27113637 Free PMC article.
-
A Benchmark for Homomeric Enzyme Active Site Structure Prediction Highlights the Importance of Accurate Modeling of Protein Symmetry.ACS Omega. 2019 Dec 19;4(27):22356-22362. doi: 10.1021/acsomega.9b02636. eCollection 2019 Dec 31. ACS Omega. 2019. PMID: 31909318 Free PMC article.
-
Construction of supramolecular nanotubes from protein crystals.Chem Sci. 2018 Oct 30;10(4):1046-1051. doi: 10.1039/c8sc04167a. eCollection 2019 Jan 28. Chem Sci. 2018. PMID: 30774900 Free PMC article.
References
-
- Howorka S. Rationally engineering natural protein assemblies in nanobiotechnology. Curr Opin Biotechnol. 2011;22:485–491. - PubMed
-
- Douglas T, Young M. Viruses: making friends with old foes. Science. 2006;312:873–875. - PubMed
-
- Lai YT, King NP, Yeates TO. Principles for designing ordered protein assemblies. Trends Cell Biol. 2012;22:653–661. - PubMed
-
- King NP, Lai YT. Practical approaches to designing novel protein assemblies. Curr Opin Struct Biol. 2013;23:632–638. - PubMed
-
- Sinclair JC. Constructing arrays of proteins. Curr Opin Chem Biol. 2013;17:946–951. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

