Large scale RNAi reveals the requirement of nuclear envelope breakdown for nuclear import of human papillomaviruses
- PMID: 24874089
- PMCID: PMC4038628
- DOI: 10.1371/journal.ppat.1004162
Large scale RNAi reveals the requirement of nuclear envelope breakdown for nuclear import of human papillomaviruses
Abstract
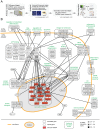
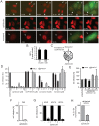
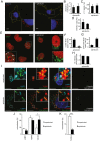
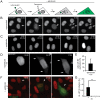
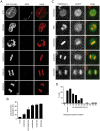
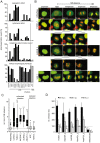
A two-step, high-throughput RNAi silencing screen was used to identify host cell factors required during human papillomavirus type 16 (HPV16) infection. Analysis of validated hits implicated a cluster of mitotic genes and revealed a previously undetermined mechanism for import of the viral DNA (vDNA) into the nucleus. In interphase cells, viruses were endocytosed, routed to the perinuclear area, and uncoated, but the vDNA failed to be imported into the nucleus. Upon nuclear envelope perforation in interphase cells HPV16 infection occured. During mitosis, the vDNA and L2 associated with host cell chromatin on the metaphase plate. Hence, we propose that HPV16 requires nuclear envelope breakdown during mitosis for access of the vDNA to the nucleoplasm. The results accentuate the value of genes found by RNAi screens for investigation of viral infections. The list of cell functions required during HPV16 infection will, moreover, provide a resource for future virus-host cell interaction studies.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
-
- Doorbar J, Quint W, Banks L, Bravo IG, Stoler M, et al. (2012) The biology and life-cycle of human papillomaviruses. Vaccine 30 Suppl 5 F55–70. - PubMed
-
- Bosch FX, de Sanjose S (2003) Chapter 1: Human papillomavirus and cervical cancer—burden and assessment of causality. J Natl Cancer Inst Monogr (31): 3–13. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

